Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
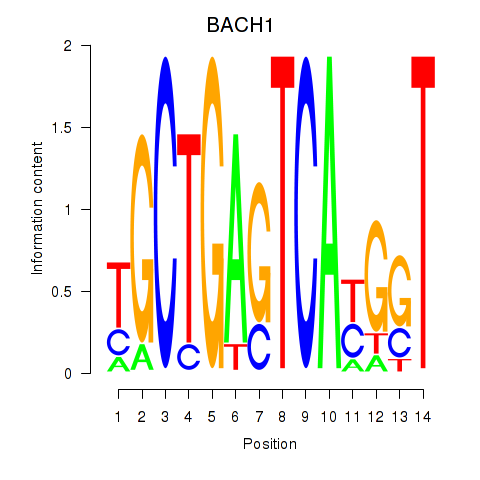
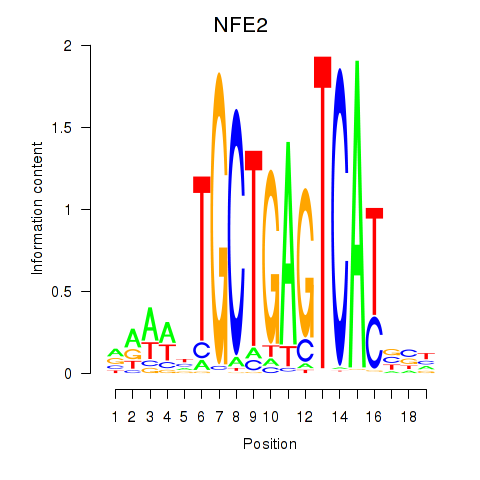
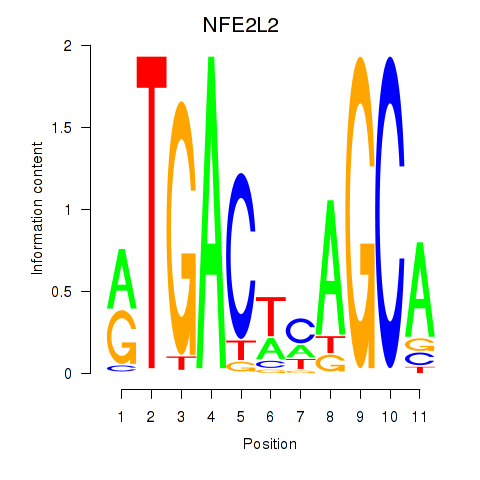
Results for BACH1_NFE2_NFE2L2
Z-value: 4.19
Transcription factors associated with BACH1_NFE2_NFE2L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH1
|
ENSG00000156273.11 | BTB domain and CNC homolog 1 |
|
NFE2
|
ENSG00000123405.9 | nuclear factor, erythroid 2 |
|
NFE2L2
|
ENSG00000116044.11 | nuclear factor, erythroid 2 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L2 | hg19_v2_chr2_-_178128199_178128245 | -0.66 | 1.5e-03 | Click! |
| BACH1 | hg19_v2_chr21_+_30671690_30671762 | -0.66 | 1.6e-03 | Click! |
| NFE2 | hg19_v2_chr12_-_54694807_54694905 | 0.07 | 7.6e-01 | Click! |
Activity profile of BACH1_NFE2_NFE2L2 motif
Sorted Z-values of BACH1_NFE2_NFE2L2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH1_NFE2_NFE2L2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.1 | GO:0016107 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 3.2 | 9.6 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 1.8 | 7.4 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 1.6 | 4.8 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 1.4 | 5.7 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 1.4 | 4.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.3 | 3.8 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 1.3 | 7.5 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 1.2 | 16.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 1.1 | 5.6 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 1.1 | 10.7 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.9 | 5.4 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.9 | 4.3 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.8 | 3.3 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.8 | 8.8 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.8 | 6.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.8 | 10.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.7 | 10.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.7 | 4.7 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.6 | 2.5 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.6 | 3.0 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.6 | 9.6 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.6 | 1.7 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.6 | 10.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.5 | 1.6 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.5 | 1.6 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.5 | 2.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.5 | 2.0 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.5 | 4.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 5.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.5 | 1.9 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.5 | 6.1 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.5 | 3.7 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.5 | 1.4 | GO:1903414 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.4 | 4.0 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.4 | 1.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 1.8 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.4 | 8.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.4 | 19.7 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.4 | 2.7 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.4 | 5.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.4 | 3.0 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.4 | 1.5 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.4 | 1.5 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.3 | 3.7 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.3 | 5.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.3 | 1.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.3 | 2.6 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.3 | 6.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 2.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.3 | 1.2 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.3 | 1.8 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.3 | 34.7 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.3 | 1.4 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.3 | 28.9 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.3 | 2.2 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.3 | 2.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.3 | 1.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.3 | 0.8 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.3 | 1.8 | GO:0060356 | leucine import(GO:0060356) |
| 0.3 | 1.8 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.3 | 1.3 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.2 | 5.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 1.4 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.2 | 0.7 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.2 | 3.4 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.2 | 0.9 | GO:0090410 | malonate catabolic process(GO:0090410) |
| 0.2 | 1.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 4.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 1.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 0.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.2 | GO:1902232 | regulation of positive thymic T cell selection(GO:1902232) |
| 0.2 | 1.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 0.9 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 0.7 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.2 | 1.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.2 | 2.7 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.2 | 1.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 7.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 0.7 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 0.5 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.2 | 2.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 1.6 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.2 | 3.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 3.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.1 | 0.5 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.5 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 1.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.6 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 6.0 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 1.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 1.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.5 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 1.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 0.8 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.6 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 1.5 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.6 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.1 | 6.7 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 1.4 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 3.5 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 3.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 3.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 4.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 2.1 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 1.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.3 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.1 | 0.4 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.1 | 2.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.5 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 1.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 3.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.7 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.5 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 1.0 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 3.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 4.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.3 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 1.4 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 1.8 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 18.0 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 2.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.4 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 1.0 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.3 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 3.0 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.4 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.4 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.3 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 2.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 1.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.0 | 10.9 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.9 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.7 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 3.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.5 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.2 | GO:0009887 | organ morphogenesis(GO:0009887) |
| 0.0 | 0.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 2.4 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 2.7 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 1.3 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.7 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 1.2 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.7 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 1.5 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 3.4 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 2.0 | GO:0007015 | actin filament organization(GO:0007015) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 1.5 | 6.1 | GO:0044753 | amphisome(GO:0044753) |
| 1.4 | 4.2 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 1.4 | 16.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.9 | 4.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.9 | 7.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.8 | 3.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.6 | 15.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.5 | 6.8 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.4 | 1.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 3.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.4 | 3.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 10.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.4 | 1.6 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.3 | 3.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 9.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 3.0 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 9.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 5.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 4.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 5.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 8.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.2 | 4.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.6 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.4 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.6 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 1.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 1.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 9.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 1.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 10.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 3.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 4.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 16.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 3.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 4.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 6.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 7.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 1.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 4.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 4.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 12.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 3.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 2.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 5.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 3.2 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 3.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 3.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 1.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 7.7 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 4.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.1 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 2.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 11.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.4 | GO:0019862 | IgA binding(GO:0019862) |
| 5.1 | 15.2 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 4.4 | 17.5 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 4.0 | 12.1 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 3.7 | 11.0 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 3.1 | 15.4 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 2.1 | 10.5 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 1.8 | 8.8 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 1.7 | 6.7 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 1.6 | 4.9 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 1.4 | 4.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 1.4 | 5.4 | GO:0004325 | ferrochelatase activity(GO:0004325) |
| 1.3 | 3.8 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 1.3 | 3.8 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.8 | 3.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.8 | 2.5 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.7 | 4.7 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.7 | 8.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.6 | 3.0 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.6 | 1.8 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.6 | 10.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.6 | 17.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.5 | 4.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.5 | 3.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.5 | 1.4 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.5 | 1.4 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.4 | 1.8 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.4 | 4.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.4 | 5.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.4 | 3.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 1.7 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.4 | 1.3 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.4 | 15.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.4 | 1.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.3 | 1.4 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.3 | 2.7 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.3 | 0.9 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.3 | 1.6 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.3 | 0.8 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.3 | 7.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 2.5 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.2 | 1.5 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.2 | 0.7 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.2 | 0.9 | GO:0090409 | malonyl-CoA synthetase activity(GO:0090409) |
| 0.2 | 4.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.2 | 1.5 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 6.0 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.2 | 1.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 5.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 3.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 2.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 1.4 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.2 | 6.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 5.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 1.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 8.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.8 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 1.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.7 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.5 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 1.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 2.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 4.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.6 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 1.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 1.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.4 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 1.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 5.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 1.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 6.2 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 1.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 9.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 3.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 13.2 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 1.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.4 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 1.0 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 7.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 3.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 3.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 3.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 2.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 3.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.5 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 2.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.6 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 9.7 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 2.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 4.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 22.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 3.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 3.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 2.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.0 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 3.1 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 5.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.5 | GO:0046966 | cytoskeletal adaptor activity(GO:0008093) thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 17.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 22.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 18.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.2 | 4.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 9.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 11.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 13.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 7.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 6.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 8.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 3.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 1.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 10.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 4.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 2.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 15.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.8 | 33.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.6 | 6.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.5 | 6.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.5 | 7.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.4 | 34.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.4 | 8.5 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.4 | 11.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 14.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 6.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 4.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 13.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 1.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 1.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 4.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 7.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 3.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 2.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 3.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 3.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 3.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 4.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 4.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 3.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 6.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 3.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.0 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 3.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 4.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 2.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |