Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
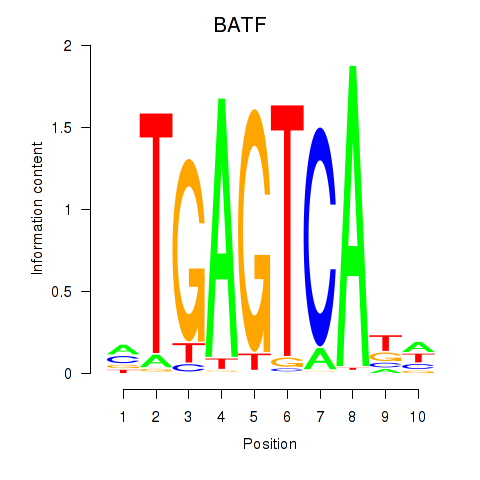
Results for BATF
Z-value: 2.49
Transcription factors associated with BATF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BATF
|
ENSG00000156127.6 | basic leucine zipper ATF-like transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BATF | hg19_v2_chr14_+_75988768_75988826 | 0.92 | 8.4e-09 | Click! |
Activity profile of BATF motif
Sorted Z-values of BATF motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BATF
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.6 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 1.7 | 5.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.3 | 5.3 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 1.3 | 4.0 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 1.3 | 11.5 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 1.1 | 7.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 1.1 | 5.4 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.0 | 3.0 | GO:1903249 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 1.0 | 3.0 | GO:0048627 | myoblast development(GO:0048627) |
| 0.9 | 4.6 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.9 | 3.7 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.9 | 9.1 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.9 | 4.5 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.8 | 2.5 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.8 | 30.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.8 | 2.4 | GO:2000449 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of T cell extravasation(GO:2000407) regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.8 | 4.7 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.8 | 2.3 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.7 | 2.9 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.7 | 4.3 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.7 | 2.0 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.7 | 0.7 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.7 | 7.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.6 | 3.0 | GO:0032796 | uropod organization(GO:0032796) |
| 0.6 | 2.3 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.6 | 2.9 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.6 | 1.7 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.5 | 5.5 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.5 | 9.7 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.5 | 4.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.5 | 1.5 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.5 | 4.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.5 | 2.0 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.5 | 1.9 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.5 | 3.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.5 | 1.4 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.5 | 11.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.4 | 2.7 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.4 | 1.8 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.4 | 1.8 | GO:0050717 | regulation of interleukin-1 alpha secretion(GO:0050705) positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.4 | 2.2 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.4 | 3.4 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.4 | 2.0 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.4 | 2.8 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.4 | 2.4 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.4 | 3.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.4 | 0.4 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.4 | 2.7 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.4 | 3.8 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.4 | 1.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.4 | 1.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.4 | 1.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.4 | 1.8 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 1.7 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 1.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 1.7 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.3 | 7.3 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.3 | 1.9 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 | 1.3 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 1.5 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.3 | 1.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.3 | 4.2 | GO:0007320 | insemination(GO:0007320) |
| 0.3 | 1.5 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.3 | 2.4 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.3 | 1.8 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.3 | 16.9 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.3 | 7.5 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.3 | 1.7 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.3 | 2.6 | GO:0002265 | astrocyte activation involved in immune response(GO:0002265) |
| 0.3 | 1.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 1.7 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.3 | 1.9 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.3 | 0.8 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.3 | 2.4 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.3 | 1.6 | GO:1900040 | regulation of interleukin-2 secretion(GO:1900040) |
| 0.3 | 2.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.3 | 1.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.3 | 1.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.3 | 0.8 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.3 | 1.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.3 | 2.4 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.3 | 1.3 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.3 | 0.3 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.3 | 5.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.3 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 1.0 | GO:1901963 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.3 | 2.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 3.0 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 1.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.2 | 1.2 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 8.2 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 1.7 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 1.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 | 1.2 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.2 | 1.4 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.2 | 1.4 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.2 | 4.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 1.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.7 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 1.5 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.2 | 0.7 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.2 | 1.9 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 3.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 2.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 1.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 3.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 0.6 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.2 | 16.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 1.0 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 | 6.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.2 | 0.2 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.2 | 0.8 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.2 | 2.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 1.9 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 1.0 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.2 | 6.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 2.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 0.8 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.2 | 3.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 0.6 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 2.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.9 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.2 | 4.8 | GO:0060004 | reflex(GO:0060004) |
| 0.2 | 0.9 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 1.8 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 | 0.4 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.2 | 1.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 1.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.2 | 1.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 1.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.2 | 0.5 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.2 | 2.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 0.9 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.2 | 1.2 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.2 | 1.0 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 0.7 | GO:0072254 | response to mycophenolic acid(GO:0071505) cellular response to mycophenolic acid(GO:0071506) metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.2 | 0.7 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 | 1.0 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.2 | 2.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 1.0 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.2 | 1.5 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.2 | 2.4 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.2 | 1.0 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 0.5 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 0.5 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.2 | 0.6 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 4.2 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.2 | 0.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.2 | 0.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 0.3 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.2 | 0.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 | 2.0 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.2 | 0.3 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.2 | 2.3 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.2 | 1.4 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.9 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.1 | 0.6 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.1 | 0.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.4 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 0.4 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 1.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 1.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.7 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 1.3 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.1 | 0.3 | GO:0046102 | inosine metabolic process(GO:0046102) |
| 0.1 | 0.9 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.1 | 0.4 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 1.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.5 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.1 | 0.4 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.1 | 10.0 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 0.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.8 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 2.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.3 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.9 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 1.6 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 1.0 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 1.3 | GO:0070777 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 1.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.5 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 1.2 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.1 | 1.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.6 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.1 | 3.7 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 0.5 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.9 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.7 | GO:0036301 | interleukin-21 production(GO:0032625) macrophage colony-stimulating factor production(GO:0036301) interleukin-21 secretion(GO:0072619) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.1 | 0.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.9 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 1.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 3.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 1.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 1.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 6.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 1.0 | GO:0001701 | in utero embryonic development(GO:0001701) |
| 0.1 | 0.2 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.1 | 0.4 | GO:0001928 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 1.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.6 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.5 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 2.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.8 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.4 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.1 | 1.0 | GO:0090197 | positive regulation of chemokine secretion(GO:0090197) |
| 0.1 | 1.4 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.1 | 0.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 1.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.6 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.7 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.3 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.1 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 2.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.8 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.4 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.0 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 2.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.4 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.1 | 0.6 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 1.8 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.1 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.1 | 0.5 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.4 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.1 | 0.9 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 2.0 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.5 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 3.8 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.3 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.8 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.6 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.2 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 1.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.2 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.7 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 1.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.1 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.1 | 0.7 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 | 0.6 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 2.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.6 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 1.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.5 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 1.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 1.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.8 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.4 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.1 | 0.5 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 3.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 2.3 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.2 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.4 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 1.7 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 5.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 2.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.8 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.4 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.4 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.6 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.1 | 1.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.4 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 2.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.2 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) positive regulation of microtubule binding(GO:1904528) |
| 0.1 | 2.4 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.2 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.1 | 1.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 0.5 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.1 | 1.7 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.1 | 1.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.4 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 1.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.2 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.1 | 5.8 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.1 | 0.5 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 0.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 0.3 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.5 | GO:0031346 | positive regulation of cell projection organization(GO:0031346) |
| 0.1 | 0.1 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.9 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 2.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.8 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 4.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.0 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 1.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.3 | GO:0018879 | biphenyl metabolic process(GO:0018879) biphenyl catabolic process(GO:0070980) |
| 0.1 | 0.2 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 1.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.2 | GO:0033319 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.1 | 0.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.8 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.9 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 1.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.3 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.1 | 1.1 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 1.8 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 4.2 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.6 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.5 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 3.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.4 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.4 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.9 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 3.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.2 | GO:1905229 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.1 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 1.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 2.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 1.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.3 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 1.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.4 | GO:0003266 | regulation of secondary heart field cardioblast proliferation(GO:0003266) |
| 0.0 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.3 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.4 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.2 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.4 | GO:0072655 | establishment of protein localization to mitochondrion(GO:0072655) |
| 0.0 | 1.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 2.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:1900044 | regulation of protein K63-linked ubiquitination(GO:1900044) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.8 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.1 | GO:0022011 | Schwann cell development(GO:0014044) myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.5 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.8 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.8 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.8 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.4 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.9 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.9 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.2 | GO:0070895 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.8 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.7 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 1.1 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 1.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 1.4 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.3 | GO:0035745 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.4 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 5.2 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.7 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.6 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.4 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.3 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.3 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.6 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.3 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 1.3 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.5 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.6 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.3 | GO:0010608 | posttranscriptional regulation of gene expression(GO:0010608) |
| 0.0 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 1.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.7 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.5 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.8 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 1.0 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 1.1 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.4 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.1 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 4.2 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 1.0 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 1.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.4 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 1.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 3.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.5 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 2.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.5 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 4.1 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.3 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.5 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 1.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.5 | GO:0090659 | walking behavior(GO:0090659) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.6 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 1.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.2 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0061209 | cell proliferation involved in mesonephros development(GO:0061209) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.6 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.3 | GO:0043623 | cellular protein complex assembly(GO:0043623) |
| 0.0 | 0.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.0 | GO:0009750 | response to fructose(GO:0009750) |
| 0.0 | 0.3 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 1.3 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.9 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.2 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) regulation of oocyte maturation(GO:1900193) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.4 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.2 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.4 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.0 | 0.2 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.0 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.4 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.4 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.3 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.7 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 1.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.0 | 1.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 1.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 21.8 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.3 | 4.0 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 1.1 | 3.4 | GO:0001534 | radial spoke(GO:0001534) |
| 0.8 | 3.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.7 | 2.0 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.7 | 4.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.7 | 2.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.6 | 1.7 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.5 | 3.1 | GO:0031673 | H zone(GO:0031673) |
| 0.5 | 3.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.5 | 1.4 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.4 | 3.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.4 | 31.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 1.6 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.4 | 1.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.4 | 1.9 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.4 | 1.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.4 | 1.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.4 | 3.5 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.3 | 6.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.3 | 2.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 1.3 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.3 | 1.5 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 1.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.3 | 1.0 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.3 | 2.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 3.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 5.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 6.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 5.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.6 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.2 | 1.6 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 1.5 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.2 | 1.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.2 | 6.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 3.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 1.0 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 0.8 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.2 | 0.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 1.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 9.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 2.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 5.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.3 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.6 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 1.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 2.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.0 | GO:0071664 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 2.6 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 5.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 1.2 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 1.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 2.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 1.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 2.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.8 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 2.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.5 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.7 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.8 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 1.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.7 | GO:0044304 | main axon(GO:0044304) |
| 0.1 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.9 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 1.7 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.9 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 10.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 8.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.5 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.4 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 2.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 2.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 2.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.5 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 4.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 4.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.2 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 2.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 2.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 0.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 5.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 5.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 3.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.9 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 6.4 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 9.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 3.0 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 7.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 18.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.9 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 3.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 4.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 1.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.0 | 1.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 10.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.2 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 3.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 3.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 1.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.5 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.8 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 1.7 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 2.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 5.7 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 1.0 | 3.0 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.9 | 6.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.9 | 5.3 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.9 | 3.4 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.8 | 2.5 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.8 | 4.2 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.8 | 2.3 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.8 | 4.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.8 | 2.3 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.7 | 7.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.6 | 1.9 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.6 | 1.9 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.6 | 13.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.6 | 7.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.6 | 5.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.6 | 3.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.6 | 2.3 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.5 | 2.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.5 | 1.5 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.5 | 1.9 | GO:0000248 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 0.5 | 2.8 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.5 | 4.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.5 | 1.4 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.5 | 1.8 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.4 | 2.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.4 | 1.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 2.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 4.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 4.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.4 | 1.8 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.3 | 1.7 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 2.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 1.3 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.3 | 5.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 2.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.3 | 1.0 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.3 | 1.9 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.3 | 1.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.3 | 5.0 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.3 | 1.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.3 | 0.9 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.3 | 4.8 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 6.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 3.8 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 3.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.7 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 3.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 2.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.2 | 3.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 1.7 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 0.9 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.2 | 1.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 0.7 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.2 | 0.7 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.2 | 3.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 9.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 7.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 0.6 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.2 | 1.0 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.2 | 0.6 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 2.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 1.3 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.2 | 0.6 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.2 | 0.8 | GO:0005115 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 2.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.2 | 2.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 2.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 0.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 5.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 1.4 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 1.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.2 | 0.7 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.2 | 6.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 1.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 3.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.7 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 2.0 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 1.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 7.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 6.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 2.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 1.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.6 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 1.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.9 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.4 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.1 | 0.9 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 2.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.1 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 1.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 3.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 19.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.5 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.5 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 1.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.8 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.6 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 0.6 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.1 | 1.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 4.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.1 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.4 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.1 | 2.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.0 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.1 | 0.4 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.4 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.5 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.8 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 2.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.7 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 2.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 2.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 5.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.9 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.9 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) arachidonic acid binding(GO:0050544) |
| 0.1 | 2.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 2.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.6 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 1.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.1 | 1.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 3.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 4.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.1 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 2.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 1.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 2.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.6 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 1.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.5 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 1.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 17.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.3 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 1.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 5.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.5 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.2 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.2 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 17.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 3.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.8 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.2 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.1 | 1.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.9 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 1.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 11.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 2.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 2.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.4 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 2.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 2.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.8 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 2.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 1.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.1 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 1.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 9.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 7.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 9.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.2 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.7 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 2.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.8 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 2.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 1.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.4 | GO:0098505 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 1.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.2 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 4.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 6.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 2.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.8 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.0 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.2 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 1.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.5 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.3 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 8.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 4.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.3 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 29.6 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0061714 | methotrexate binding(GO:0051870) folic acid receptor activity(GO:0061714) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.0 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.3 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 1.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.0 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 1.5 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.1 | GO:0050610 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.3 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) |
| 0.0 | 0.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 6.7 | GO:0005198 | structural molecule activity(GO:0005198) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 28.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 11.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 0.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 9.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 0.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 2.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 3.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 6.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 8.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 3.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 8.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 6.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 0.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 1.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 1.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 8.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 2.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 4.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 3.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 4.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 4.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.2 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 33.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 4.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 4.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 3.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 7.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 4.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 5.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 5.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 10.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 5.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 3.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.1 | 1.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 3.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 3.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.6 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.5 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 2.3 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 1.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 3.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 14.1 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.8 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.6 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 2.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 5.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |