Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
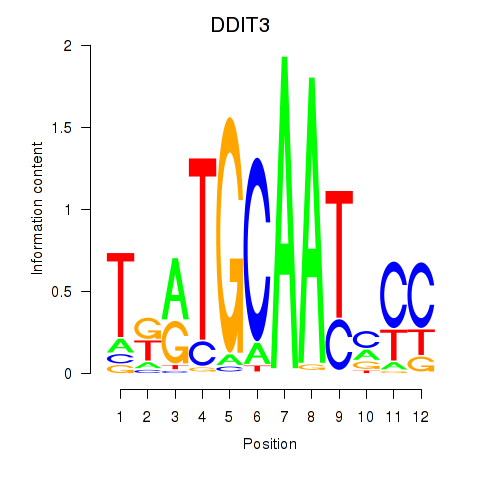
Results for DDIT3
Z-value: 1.51
Transcription factors associated with DDIT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DDIT3
|
ENSG00000175197.6 | DNA damage inducible transcript 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DDIT3 | hg19_v2_chr12_-_57914275_57914304 | -0.41 | 7.4e-02 | Click! |
Activity profile of DDIT3 motif
Sorted Z-values of DDIT3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DDIT3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.6 | 2.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.4 | 2.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.3 | 1.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.3 | 2.0 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.3 | 1.0 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.3 | 3.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 1.4 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 2.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 0.8 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.4 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.8 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.8 | GO:0031860 | telomeric loop formation(GO:0031627) telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 1.0 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.9 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 0.8 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.1 | 0.6 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 1.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 1.2 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.6 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 0.3 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 2.6 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.1 | 0.6 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 0.9 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 1.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 2.6 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 0.9 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 2.7 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.7 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.3 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.8 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 2.0 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.3 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.2 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.4 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 1.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.4 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 3.7 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 4.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0071942 | XPC complex(GO:0071942) |
| 0.3 | 2.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 2.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.6 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 2.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.3 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.1 | 0.3 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.9 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.3 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 0.0 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 6.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 2.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.6 | 2.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.2 | 0.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.4 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 1.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 0.8 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 2.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 1.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 3.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.9 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 2.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.7 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 2.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 1.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.8 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 2.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 3.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 2.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) virion binding(GO:0046790) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 1.2 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.4 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 3.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 2.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 2.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |