Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
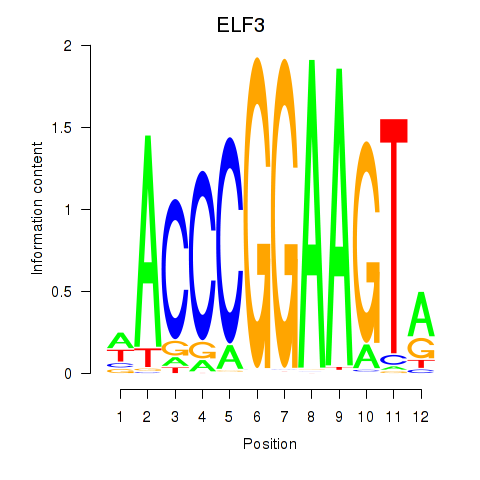
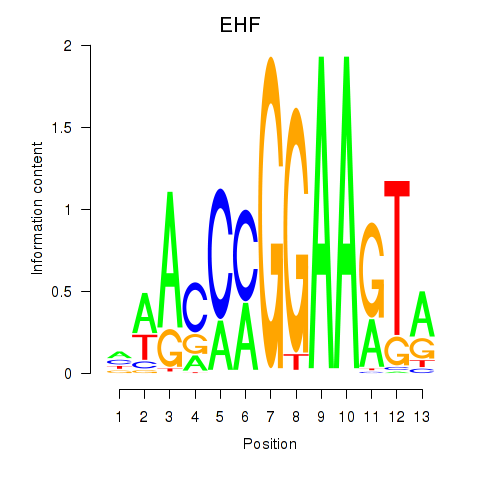
Results for ELF3_EHF
Z-value: 3.91
Transcription factors associated with ELF3_EHF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ELF3
|
ENSG00000163435.11 | E74 like ETS transcription factor 3 |
|
EHF
|
ENSG00000135373.8 | ETS homologous factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EHF | hg19_v2_chr11_+_34643600_34643750 | 0.95 | 6.8e-11 | Click! |
| ELF3 | hg19_v2_chr1_+_201979743_201979772 | -0.81 | 1.3e-05 | Click! |
Activity profile of ELF3_EHF motif
Sorted Z-values of ELF3_EHF motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ELF3_EHF
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 18.7 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 2.0 | 6.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.9 | 5.6 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 1.8 | 18.3 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 1.6 | 9.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 1.4 | 1.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 1.2 | 7.0 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 1.2 | 5.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.1 | 4.5 | GO:0016333 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 1.1 | 4.4 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 1.1 | 3.2 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 1.1 | 6.4 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 1.0 | 5.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 1.0 | 2.9 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 1.0 | 5.8 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.9 | 2.8 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.9 | 5.7 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.9 | 2.7 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.9 | 2.6 | GO:2001226 | negative regulation of anion channel activity(GO:0010360) negative regulation of chloride transport(GO:2001226) |
| 0.8 | 3.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.8 | 4.9 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.8 | 2.4 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.7 | 4.4 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.7 | 2.1 | GO:0043315 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.7 | 3.5 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.7 | 7.6 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.7 | 1.4 | GO:0043217 | peripheral nervous system myelin maintenance(GO:0032287) myelin maintenance(GO:0043217) |
| 0.7 | 6.9 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.7 | 3.4 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.6 | 1.9 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.6 | 4.8 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.6 | 0.6 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.6 | 1.7 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.6 | 0.6 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.5 | 1.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.5 | 2.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.5 | 1.1 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.5 | 4.0 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.5 | 2.4 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.5 | 1.9 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.5 | 3.8 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.5 | 1.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.4 | 4.0 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.4 | 2.7 | GO:0042335 | cuticle development(GO:0042335) |
| 0.4 | 7.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.4 | 3.1 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.4 | 5.2 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.4 | 7.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.4 | 2.6 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.4 | 3.0 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.4 | 2.9 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.4 | 7.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.4 | 1.2 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.4 | 1.2 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.4 | 1.2 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.4 | 9.0 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.4 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.4 | 1.5 | GO:0048850 | chemoattraction of serotonergic neuron axon(GO:0036517) hypophysis morphogenesis(GO:0048850) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.4 | 1.9 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.4 | 8.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.4 | 2.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.4 | 1.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.4 | 2.8 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.3 | 1.4 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.3 | 2.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.3 | 1.4 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.3 | 1.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.3 | 1.4 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.3 | 0.3 | GO:0030168 | platelet activation(GO:0030168) |
| 0.3 | 1.7 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.3 | 2.3 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.3 | 6.0 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.3 | 2.6 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.3 | 1.9 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.3 | 6.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 | 1.6 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.3 | 0.9 | GO:1902309 | regulation of heart rate by hormone(GO:0003064) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.3 | 2.8 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 5.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 5.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.3 | 3.3 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.3 | 0.9 | GO:1901253 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.3 | 3.6 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.3 | 2.4 | GO:1904382 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.3 | 1.5 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.3 | 0.9 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.3 | 4.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 0.8 | GO:0002276 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.3 | 3.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.3 | 14.2 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.3 | 0.3 | GO:0072387 | flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.3 | 0.5 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.3 | 2.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.3 | 2.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 1.0 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.3 | 0.5 | GO:0031297 | replication fork processing(GO:0031297) DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.3 | 5.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.2 | 1.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.2 | 2.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 0.7 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.2 | 5.4 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.2 | 0.7 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.2 | 0.9 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.2 | 1.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 4.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 0.7 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.2 | 4.4 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.2 | 0.9 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.2 | 1.3 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 | 1.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.2 | 2.8 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.2 | 0.4 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.2 | 3.7 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.2 | 2.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 0.6 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.2 | 3.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 2.4 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 0.8 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 0.6 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.2 | 1.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 0.8 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.2 | 11.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 0.5 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.2 | 0.5 | GO:1902824 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.2 | 0.9 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.2 | 1.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 0.4 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.2 | 0.9 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 0.5 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.2 | 3.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 0.7 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.2 | 2.0 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.2 | 0.5 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.2 | 5.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 2.0 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 1.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 0.5 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.2 | 2.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.2 | 0.3 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.2 | 0.5 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.2 | 6.3 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.2 | 0.6 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.2 | 0.9 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.9 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.2 | 0.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 0.9 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.2 | 0.8 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.2 | 2.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.6 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.1 | 0.6 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.7 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.4 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.1 | 12.3 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 0.4 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 5.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.3 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 2.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.6 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 2.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 0.7 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 2.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 6.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.4 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 | 1.2 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 1.6 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.1 | 0.6 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 1.9 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 2.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.1 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 1.7 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 1.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 3.8 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.1 | 1.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 1.4 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 3.6 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 5.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.8 | GO:0044782 | cilium organization(GO:0044782) |
| 0.1 | 1.9 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.5 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.6 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 1.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 6.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.5 | GO:0034101 | erythrocyte homeostasis(GO:0034101) |
| 0.1 | 1.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.0 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.1 | 0.6 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.3 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.1 | 0.3 | GO:0070781 | response to biotin(GO:0070781) |
| 0.1 | 1.4 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 | 0.5 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 7.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.6 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.5 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.1 | 0.3 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 1.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 0.9 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.4 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 0.7 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 1.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 7.6 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.3 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.7 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.1 | 1.1 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 0.9 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.4 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 2.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 1.6 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.6 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 1.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 0.6 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 5.3 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 13.2 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.1 | 1.9 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.8 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 1.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 4.0 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 0.6 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 2.6 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 0.6 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 4.5 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.1 | 1.5 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 1.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.5 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.5 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 2.5 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 1.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 3.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 1.0 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 1.3 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.4 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 1.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 3.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.9 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 0.9 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.1 | 0.3 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.1 | 0.6 | GO:1900086 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 0.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 1.2 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 4.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 2.2 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 1.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.6 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.3 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) |
| 0.1 | 1.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.2 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 1.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 0.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.3 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.2 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 6.4 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 1.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.3 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 0.4 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.5 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 1.4 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.1 | 1.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.4 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.1 | 1.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 1.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 3.9 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 4.6 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.6 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 2.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 1.9 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 1.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.6 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 1.0 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 0.4 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.5 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.1 | GO:0043307 | eosinophil activation(GO:0043307) |
| 0.1 | 1.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 1.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 1.3 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.7 | GO:1902186 | viral release from host cell(GO:0019076) regulation of viral release from host cell(GO:1902186) |
| 0.0 | 0.1 | GO:0061724 | lipophagy(GO:0061724) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 1.0 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.7 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 1.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.7 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.7 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.2 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.5 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 1.6 | GO:0006366 | transcription from RNA polymerase II promoter(GO:0006366) |
| 0.0 | 0.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 6.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 2.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0036093 | male germ cell proliferation(GO:0002176) germ cell proliferation(GO:0036093) |
| 0.0 | 0.7 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 1.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.9 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.0 | 0.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.9 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.4 | GO:0061430 | bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.7 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.2 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.4 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 1.6 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.2 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.0 | 3.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.5 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.4 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 1.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:0030193 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 0.2 | GO:0002396 | MHC protein complex assembly(GO:0002396) MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.9 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.4 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.4 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 1.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.3 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.5 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.7 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 3.0 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 1.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.6 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.4 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.0 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.9 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.3 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 1.3 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 1.0 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.7 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 2.0 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 0.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) |
| 0.0 | 0.2 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 1.2 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.6 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.7 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 0.3 | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 5.9 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.5 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.0 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.5 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.5 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.0 | GO:0090410 | malonate catabolic process(GO:0090410) |
| 0.0 | 0.4 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.3 | GO:0033619 | membrane protein proteolysis(GO:0033619) |
| 0.0 | 0.3 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 2.3 | 18.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.6 | 12.4 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 1.5 | 18.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 1.0 | 4.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.9 | 4.5 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.9 | 2.6 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.9 | 5.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.8 | 5.0 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.8 | 9.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.7 | 0.7 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.6 | 4.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.5 | 3.8 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.5 | 2.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.5 | 2.0 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.5 | 2.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.5 | 0.9 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.5 | 1.8 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.4 | 1.3 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.4 | 0.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.4 | 2.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.3 | 1.7 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 2.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 11.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.3 | 1.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.3 | 8.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.3 | 3.0 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.3 | 10.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.3 | 1.7 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.3 | 1.3 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.3 | 4.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 0.9 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.3 | 2.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 3.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.3 | 2.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.3 | 1.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.3 | 1.6 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.3 | 1.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.3 | 2.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 3.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 2.5 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 1.9 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 1.0 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.2 | 2.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 1.5 | GO:0034448 | EGO complex(GO:0034448) |
| 0.2 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 1.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 1.0 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.2 | 1.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 3.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 2.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 2.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 1.9 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 1.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.2 | 1.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 1.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 2.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 1.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 0.6 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 0.5 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 16.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 3.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 2.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 0.6 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.1 | 9.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 13.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 3.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.5 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 1.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 3.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 3.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 5.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 1.0 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 1.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 1.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.8 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 0.7 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.9 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 6.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.5 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 1.0 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.1 | 2.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 13.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) glial limiting end-foot(GO:0097451) |
| 0.1 | 7.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.7 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.1 | 1.0 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 14.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 4.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 1.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.3 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 1.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 2.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 3.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 1.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 3.0 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 3.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 6.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 10.4 | GO:0070160 | occluding junction(GO:0070160) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 7.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.3 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.3 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 4.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.1 | 0.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.8 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 3.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 1.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.8 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 1.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 3.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 4.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.5 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.3 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 3.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.7 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.4 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.7 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 1.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 2.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.4 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 21.5 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 2.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 3.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0044433 | cytoplasmic vesicle part(GO:0044433) |
| 0.0 | 1.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.0 | GO:0098837 | insulin-responsive compartment(GO:0032593) postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.9 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 1.0 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 12.4 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 1.4 | 6.9 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 1.1 | 5.6 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.9 | 3.5 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.8 | 3.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.8 | 7.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.8 | 18.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.7 | 2.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.7 | 2.7 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.6 | 1.7 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.5 | 3.8 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.5 | 3.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.5 | 3.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.5 | 1.5 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.5 | 12.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.5 | 1.9 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.5 | 2.9 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.5 | 3.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.5 | 3.8 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.5 | 1.9 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.5 | 7.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.5 | 18.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 1.9 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.5 | 1.4 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.4 | 4.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.4 | 2.6 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.4 | 0.8 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.4 | 4.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 1.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 1.5 | GO:0005115 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.4 | 16.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.4 | 5.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 1.7 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.3 | 4.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 6.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 1.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.3 | 4.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.3 | 2.9 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 0.9 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.3 | 2.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.3 | 1.2 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.3 | 1.5 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.3 | 1.8 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.3 | 1.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 1.7 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.3 | 2.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.3 | 4.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 3.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 7.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 2.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 2.0 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 5.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 4.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 3.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 1.4 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 4.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 5.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 13.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 3.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 2.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.2 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.2 | 1.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 0.9 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.2 | 5.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 1.3 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.2 | 1.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 1.7 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.2 | 0.6 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 1.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 4.8 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 0.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 0.8 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 0.6 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 1.4 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.2 | 2.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 6.0 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.2 | 11.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.2 | 0.9 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.2 | 2.6 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 0.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 0.5 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 1.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 0.7 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 0.5 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.2 | 0.8 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.2 | 5.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 5.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 7.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 4.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 0.5 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.2 | 3.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 2.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 0.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 3.8 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 0.5 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.2 | 0.6 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.2 | 2.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.2 | 2.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 1.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 0.6 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 0.7 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.4 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.7 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 1.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.4 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 4.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.9 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 2.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 2.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.9 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.7 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 8.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 2.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.3 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.1 | 6.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.5 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 4.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.3 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 1.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 2.5 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 1.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 2.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 2.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 11.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 4.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 4.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.9 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 18.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 2.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.6 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 1.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 2.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 1.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 2.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 2.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 3.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 20.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 6.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 3.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 2.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 3.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 6.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 2.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 4.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.9 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.5 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 1.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 2.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 1.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 8.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 3.2 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.1 | 1.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.4 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 1.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 89.8 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 5.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0033743 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 1.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.6 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.9 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 1.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 1.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.6 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 15.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.4 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 6.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.2 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0035586 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) purinergic receptor activity(GO:0035586) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 10.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 1.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.0 | GO:0090409 | malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 8.7 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 18.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 26.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.4 | 3.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 9.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 6.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 10.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 3.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.9 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 7.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 13.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 8.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 7.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 4.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 5.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 9.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 5.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 1.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 4.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 5.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 3.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 8.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 2.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 6.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 2.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 15.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.5 | 12.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 13.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.3 | 4.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 3.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 3.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 6.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 4.8 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.2 | 4.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 6.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 7.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 9.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 5.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 6.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 4.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 3.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 8.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 5.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 3.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 7.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.3 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 1.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 4.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 4.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 5.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 4.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 11.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 19.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 42.4 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 1.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.3 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 8.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 4.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 4.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 2.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 3.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 2.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 4.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 3.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 4.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |