Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
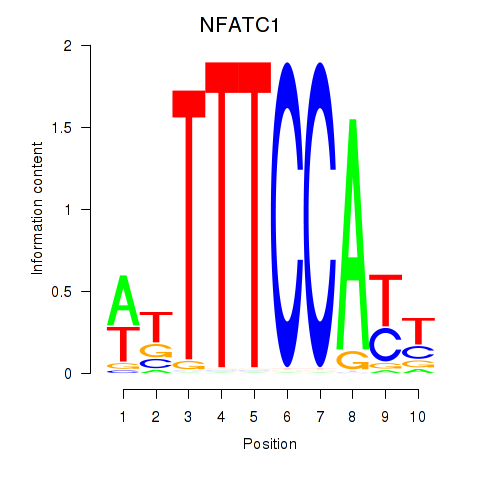
Results for NFATC1
Z-value: 0.82
Transcription factors associated with NFATC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC1
|
ENSG00000131196.18 | nuclear factor of activated T cells 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC1 | hg38_v1_chr18_+_79395856_79395939 | -0.61 | 1.1e-01 | Click! |
Activity profile of NFATC1 motif
Sorted Z-values of NFATC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_91946989 | 2.29 |
ENST00000556154.5
|
FBLN5
|
fibulin 5 |
| chr14_-_91947383 | 2.02 |
ENST00000267620.14
|
FBLN5
|
fibulin 5 |
| chr2_+_188974364 | 1.29 |
ENST00000304636.9
ENST00000317840.9 |
COL3A1
|
collagen type III alpha 1 chain |
| chr12_-_91179472 | 1.06 |
ENST00000550099.5
ENST00000546391.5 |
DCN
|
decorin |
| chr12_-_91179355 | 1.03 |
ENST00000550563.5
ENST00000546370.5 |
DCN
|
decorin |
| chr7_+_100602344 | 0.68 |
ENST00000223061.6
|
PCOLCE
|
procollagen C-endopeptidase enhancer |
| chr15_+_32717994 | 0.64 |
ENST00000560677.5
ENST00000560830.1 ENST00000651154.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chrX_+_153494970 | 0.62 |
ENST00000331595.9
ENST00000431891.1 |
BGN
|
biglycan |
| chr13_-_67230313 | 0.59 |
ENST00000377865.7
|
PCDH9
|
protocadherin 9 |
| chr18_-_28036585 | 0.59 |
ENST00000399380.7
|
CDH2
|
cadherin 2 |
| chr13_-_67230377 | 0.55 |
ENST00000544246.5
ENST00000377861.4 |
PCDH9
|
protocadherin 9 |
| chr1_+_162632454 | 0.53 |
ENST00000367921.8
ENST00000367922.7 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr2_-_162242998 | 0.50 |
ENST00000627638.2
ENST00000447386.5 |
FAP
|
fibroblast activation protein alpha |
| chr5_-_111758061 | 0.46 |
ENST00000509979.5
ENST00000513100.5 ENST00000508161.5 ENST00000455559.6 |
NREP
|
neuronal regeneration related protein |
| chr8_-_13514821 | 0.45 |
ENST00000276297.9
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr1_-_153544997 | 0.44 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4 |
| chr3_+_148730100 | 0.44 |
ENST00000474935.5
ENST00000475347.5 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor type 1 |
| chr5_-_159099684 | 0.41 |
ENST00000380654.8
|
EBF1
|
EBF transcription factor 1 |
| chr12_+_78863962 | 0.41 |
ENST00000393240.7
|
SYT1
|
synaptotagmin 1 |
| chr5_-_159099745 | 0.41 |
ENST00000517373.1
|
EBF1
|
EBF transcription factor 1 |
| chr17_-_15260752 | 0.40 |
ENST00000676329.1
ENST00000675551.1 ENST00000644020.1 ENST00000674947.1 |
PMP22
|
peripheral myelin protein 22 |
| chr5_-_159099909 | 0.39 |
ENST00000313708.11
|
EBF1
|
EBF transcription factor 1 |
| chr9_+_88991440 | 0.38 |
ENST00000358157.3
|
S1PR3
|
sphingosine-1-phosphate receptor 3 |
| chr8_-_13514744 | 0.38 |
ENST00000316609.9
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr3_-_71583683 | 0.36 |
ENST00000649631.1
ENST00000648718.1 |
FOXP1
|
forkhead box P1 |
| chr9_-_16870702 | 0.34 |
ENST00000380667.6
ENST00000545497.5 ENST00000486514.5 |
BNC2
|
basonuclin 2 |
| chr10_+_92831153 | 0.34 |
ENST00000672817.1
|
EXOC6
|
exocyst complex component 6 |
| chr10_-_103452384 | 0.34 |
ENST00000369788.7
|
CALHM2
|
calcium homeostasis modulator family member 2 |
| chr8_+_76681208 | 0.34 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr16_+_15434577 | 0.34 |
ENST00000300006.9
|
BMERB1
|
bMERB domain containing 1 |
| chr16_+_15434475 | 0.34 |
ENST00000566490.5
|
BMERB1
|
bMERB domain containing 1 |
| chr10_+_103277129 | 0.33 |
ENST00000369849.9
|
INA
|
internexin neuronal intermediate filament protein alpha |
| chr6_-_56851888 | 0.32 |
ENST00000312431.10
ENST00000520645.5 |
DST
|
dystonin |
| chr10_-_103452356 | 0.32 |
ENST00000260743.10
|
CALHM2
|
calcium homeostasis modulator family member 2 |
| chr3_-_15797930 | 0.30 |
ENST00000683139.1
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr11_-_102530738 | 0.30 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7 |
| chr1_+_78649818 | 0.29 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr3_-_71583713 | 0.29 |
ENST00000649528.3
ENST00000471386.3 ENST00000493089.7 |
FOXP1
|
forkhead box P1 |
| chr2_-_213151590 | 0.28 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr8_-_121641424 | 0.27 |
ENST00000303924.5
|
HAS2
|
hyaluronan synthase 2 |
| chr5_+_141350081 | 0.27 |
ENST00000523390.2
ENST00000611598.1 |
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr9_-_20622479 | 0.25 |
ENST00000380338.9
|
MLLT3
|
MLLT3 super elongation complex subunit |
| chr2_-_162243375 | 0.24 |
ENST00000188790.9
ENST00000443424.5 |
FAP
|
fibroblast activation protein alpha |
| chr7_+_120988683 | 0.23 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr5_+_149141483 | 0.23 |
ENST00000326685.11
ENST00000309868.12 |
ABLIM3
|
actin binding LIM protein family member 3 |
| chr5_+_149141573 | 0.22 |
ENST00000506113.5
|
ABLIM3
|
actin binding LIM protein family member 3 |
| chr3_-_71583592 | 0.22 |
ENST00000650156.1
ENST00000649596.1 |
FOXP1
|
forkhead box P1 |
| chr7_-_29146436 | 0.22 |
ENST00000396276.7
|
CPVL
|
carboxypeptidase vitellogenic like |
| chr3_+_69763726 | 0.21 |
ENST00000448226.9
|
MITF
|
melanocyte inducing transcription factor |
| chr15_+_92900189 | 0.21 |
ENST00000626874.2
ENST00000627622.1 ENST00000629346.2 ENST00000628375.2 ENST00000420239.7 ENST00000394196.9 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr8_-_130386864 | 0.21 |
ENST00000521426.5
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chrX_+_22136552 | 0.20 |
ENST00000682888.1
ENST00000684356.1 |
PHEX
|
phosphate regulating endopeptidase homolog X-linked |
| chr8_+_38974212 | 0.19 |
ENST00000302495.5
|
HTRA4
|
HtrA serine peptidase 4 |
| chr17_-_31321603 | 0.19 |
ENST00000462804.3
|
EVI2A
|
ecotropic viral integration site 2A |
| chr12_-_10453330 | 0.19 |
ENST00000347831.9
ENST00000359151.8 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr3_-_127822455 | 0.19 |
ENST00000265052.10
|
MGLL
|
monoglyceride lipase |
| chr4_-_158173042 | 0.18 |
ENST00000592057.1
ENST00000393807.9 |
GASK1B
|
golgi associated kinase 1B |
| chr17_-_31321743 | 0.18 |
ENST00000247270.3
|
EVI2A
|
ecotropic viral integration site 2A |
| chr3_-_115147237 | 0.18 |
ENST00000357258.8
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr9_-_70869076 | 0.18 |
ENST00000677594.1
|
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr3_-_115147277 | 0.18 |
ENST00000675478.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr4_-_158173004 | 0.17 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B |
| chr5_-_160685379 | 0.17 |
ENST00000642502.1
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr1_+_85580751 | 0.17 |
ENST00000451137.7
|
CCN1
|
cellular communication network factor 1 |
| chr4_+_122339221 | 0.17 |
ENST00000442707.1
|
KIAA1109
|
KIAA1109 |
| chr12_+_1820558 | 0.17 |
ENST00000299194.6
|
LRTM2
|
leucine rich repeats and transmembrane domains 2 |
| chr12_+_1820255 | 0.17 |
ENST00000543818.5
|
LRTM2
|
leucine rich repeats and transmembrane domains 2 |
| chr16_-_18876305 | 0.17 |
ENST00000563235.5
|
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chrX_-_100410264 | 0.16 |
ENST00000373034.8
|
PCDH19
|
protocadherin 19 |
| chr14_+_104581141 | 0.16 |
ENST00000410013.1
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr4_-_41748713 | 0.16 |
ENST00000226382.4
|
PHOX2B
|
paired like homeobox 2B |
| chr11_-_58844695 | 0.16 |
ENST00000287275.6
|
GLYATL2
|
glycine-N-acyltransferase like 2 |
| chrX_-_63785510 | 0.16 |
ENST00000437457.6
ENST00000374878.5 ENST00000623517.3 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chr3_+_69084929 | 0.16 |
ENST00000273258.4
|
ARL6IP5
|
ADP ribosylation factor like GTPase 6 interacting protein 5 |
| chr12_-_91179517 | 0.16 |
ENST00000551354.1
|
DCN
|
decorin |
| chr4_-_177442427 | 0.16 |
ENST00000264595.7
|
AGA
|
aspartylglucosaminidase |
| chr12_+_1820617 | 0.16 |
ENST00000535041.5
|
LRTM2
|
leucine rich repeats and transmembrane domains 2 |
| chr3_-_157533594 | 0.16 |
ENST00000487753.5
ENST00000489602.1 ENST00000461299.5 ENST00000479987.5 |
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr3_+_152300135 | 0.15 |
ENST00000465907.6
ENST00000492948.5 ENST00000485509.5 ENST00000464596.5 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr7_-_100896123 | 0.15 |
ENST00000428317.7
|
ACHE
|
acetylcholinesterase (Cartwright blood group) |
| chr10_-_78029487 | 0.15 |
ENST00000372371.8
|
POLR3A
|
RNA polymerase III subunit A |
| chr2_+_33476640 | 0.15 |
ENST00000425210.5
ENST00000444784.5 ENST00000423159.5 ENST00000403687.8 |
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr2_+_11677587 | 0.15 |
ENST00000449576.6
|
LPIN1
|
lipin 1 |
| chr14_+_50560137 | 0.15 |
ENST00000358385.12
|
ATL1
|
atlastin GTPase 1 |
| chr5_-_55173173 | 0.15 |
ENST00000296733.5
ENST00000322374.10 ENST00000381375.7 |
CDC20B
|
cell division cycle 20B |
| chr9_-_70869029 | 0.14 |
ENST00000361823.9
ENST00000377101.5 ENST00000360823.6 ENST00000377105.5 |
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr19_-_43883964 | 0.14 |
ENST00000587539.2
|
ZNF404
|
zinc finger protein 404 |
| chr22_+_37675629 | 0.14 |
ENST00000215909.10
|
LGALS1
|
galectin 1 |
| chr5_+_50666612 | 0.13 |
ENST00000281631.10
|
PARP8
|
poly(ADP-ribose) polymerase family member 8 |
| chr12_+_25958891 | 0.13 |
ENST00000381352.7
ENST00000535907.5 ENST00000405154.6 ENST00000615708.4 |
RASSF8
|
Ras association domain family member 8 |
| chr4_-_16083714 | 0.13 |
ENST00000508167.5
|
PROM1
|
prominin 1 |
| chr3_+_69084973 | 0.13 |
ENST00000478935.1
|
ARL6IP5
|
ADP ribosylation factor like GTPase 6 interacting protein 5 |
| chr5_+_71719267 | 0.13 |
ENST00000296777.5
|
CARTPT
|
CART prepropeptide |
| chr10_-_102837406 | 0.13 |
ENST00000369887.4
ENST00000638272.1 ENST00000639393.1 ENST00000638971.1 ENST00000638190.1 |
CYP17A1
|
cytochrome P450 family 17 subfamily A member 1 |
| chr5_+_140401808 | 0.13 |
ENST00000616482.4
ENST00000297183.10 ENST00000360839.7 ENST00000421134.5 ENST00000394723.7 ENST00000511151.5 |
ANKHD1
|
ankyrin repeat and KH domain containing 1 |
| chr4_-_16083695 | 0.13 |
ENST00000510224.5
|
PROM1
|
prominin 1 |
| chr4_+_26584064 | 0.12 |
ENST00000264866.9
ENST00000505206.5 ENST00000511789.5 |
TBC1D19
|
TBC1 domain family member 19 |
| chr5_-_74641419 | 0.12 |
ENST00000618628.4
ENST00000510316.5 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 |
| chr1_-_150808251 | 0.12 |
ENST00000271651.8
ENST00000676970.1 ENST00000679260.1 ENST00000676751.1 ENST00000677887.1 |
CTSK
|
cathepsin K |
| chr7_-_120858066 | 0.12 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr14_+_80955577 | 0.12 |
ENST00000642209.1
ENST00000298171.7 |
TSHR
|
thyroid stimulating hormone receptor |
| chr10_-_63269057 | 0.12 |
ENST00000542921.5
|
JMJD1C
|
jumonji domain containing 1C |
| chr16_+_1533654 | 0.12 |
ENST00000566264.2
|
TMEM204
|
transmembrane protein 204 |
| chr16_-_21425278 | 0.12 |
ENST00000504841.6
ENST00000419180.6 |
NPIPB3
|
nuclear pore complex interacting protein family member B3 |
| chr4_-_82844418 | 0.12 |
ENST00000503937.5
|
SEC31A
|
SEC31 homolog A, COPII coat complex component |
| chrX_+_41333905 | 0.12 |
ENST00000457138.7
ENST00000643821.1 |
DDX3X
|
DEAD-box helicase 3 X-linked |
| chr2_-_40452046 | 0.12 |
ENST00000406785.6
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr6_+_10528326 | 0.12 |
ENST00000379597.7
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr7_+_134866831 | 0.12 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr14_-_89417148 | 0.12 |
ENST00000557258.6
|
FOXN3
|
forkhead box N3 |
| chrX_-_13817346 | 0.12 |
ENST00000356942.9
|
GPM6B
|
glycoprotein M6B |
| chr3_-_15798184 | 0.12 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr10_-_13099652 | 0.12 |
ENST00000378839.1
|
CCDC3
|
coiled-coil domain containing 3 |
| chr3_-_179259208 | 0.12 |
ENST00000485523.5
|
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr2_-_55334529 | 0.11 |
ENST00000645860.1
ENST00000642563.1 ENST00000647396.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr10_-_90921079 | 0.11 |
ENST00000371697.4
|
ANKRD1
|
ankyrin repeat domain 1 |
| chr11_-_2161158 | 0.11 |
ENST00000421783.1
ENST00000397262.5 ENST00000381330.5 ENST00000250971.7 ENST00000397270.1 |
INS
INS-IGF2
|
insulin INS-IGF2 readthrough |
| chr1_+_61082702 | 0.11 |
ENST00000485903.6
ENST00000371185.6 ENST00000371184.6 |
NFIA
|
nuclear factor I A |
| chrX_+_9560465 | 0.11 |
ENST00000647060.1
|
TBL1X
|
transducin beta like 1 X-linked |
| chr15_-_43493076 | 0.11 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr7_+_20647388 | 0.11 |
ENST00000258738.10
|
ABCB5
|
ATP binding cassette subfamily B member 5 |
| chr1_-_27490045 | 0.11 |
ENST00000536657.1
|
WASF2
|
WASP family member 2 |
| chr1_-_154192058 | 0.11 |
ENST00000271850.11
ENST00000368530.7 ENST00000651641.1 |
TPM3
|
tropomyosin 3 |
| chr14_+_58427970 | 0.11 |
ENST00000261244.9
|
KIAA0586
|
KIAA0586 |
| chr6_+_32439866 | 0.11 |
ENST00000374982.5
ENST00000395388.7 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr10_+_102776237 | 0.11 |
ENST00000369889.5
|
WBP1L
|
WW domain binding protein 1 like |
| chr6_-_135498417 | 0.11 |
ENST00000681022.1
ENST00000680033.1 |
AHI1
|
Abelson helper integration site 1 |
| chr2_+_46699241 | 0.11 |
ENST00000394861.3
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr17_+_34255274 | 0.11 |
ENST00000580907.5
ENST00000225831.4 |
CCL2
|
C-C motif chemokine ligand 2 |
| chr15_-_43493105 | 0.11 |
ENST00000382039.7
ENST00000450115.6 ENST00000382044.9 |
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr1_+_15476099 | 0.10 |
ENST00000375910.8
|
CELA2B
|
chymotrypsin like elastase 2B |
| chr8_+_84183534 | 0.10 |
ENST00000518566.5
|
RALYL
|
RALY RNA binding protein like |
| chr6_-_79234713 | 0.10 |
ENST00000620514.1
|
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr6_-_79234619 | 0.10 |
ENST00000344726.9
ENST00000275036.11 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr9_+_35161951 | 0.10 |
ENST00000617908.4
ENST00000619578.4 |
UNC13B
|
unc-13 homolog B |
| chr12_+_26195543 | 0.10 |
ENST00000242729.7
|
SSPN
|
sarcospan |
| chr17_+_7425606 | 0.10 |
ENST00000333870.8
ENST00000574034.1 |
SPEM2
|
SPEM family member 2 |
| chr9_-_35815034 | 0.10 |
ENST00000259667.6
|
HINT2
|
histidine triad nucleotide binding protein 2 |
| chr9_+_121299793 | 0.10 |
ENST00000373818.8
|
GSN
|
gelsolin |
| chr4_+_41613476 | 0.10 |
ENST00000508466.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr17_+_81528370 | 0.10 |
ENST00000417245.7
ENST00000334850.7 |
FSCN2
|
fascin actin-bundling protein 2, retinal |
| chr9_+_35162000 | 0.10 |
ENST00000396787.5
ENST00000378495.7 ENST00000635942.1 ENST00000378496.8 |
UNC13B
|
unc-13 homolog B |
| chr1_-_41662298 | 0.09 |
ENST00000643665.1
|
HIVEP3
|
HIVEP zinc finger 3 |
| chr19_-_18281612 | 0.09 |
ENST00000252818.5
|
JUND
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr3_-_179266971 | 0.09 |
ENST00000349697.2
ENST00000497599.5 |
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr5_+_140401860 | 0.09 |
ENST00000532219.5
ENST00000394722.7 |
ANKHD1-EIF4EBP3
ANKHD1
|
ANKHD1-EIF4EBP3 readthrough ankyrin repeat and KH domain containing 1 |
| chr19_-_14062028 | 0.09 |
ENST00000669674.2
|
PALM3
|
paralemmin 3 |
| chr2_+_200526120 | 0.09 |
ENST00000409203.3
ENST00000357799.9 |
SGO2
|
shugoshin 2 |
| chr11_+_118883884 | 0.09 |
ENST00000292174.5
|
CXCR5
|
C-X-C motif chemokine receptor 5 |
| chr16_+_28638065 | 0.09 |
ENST00000683297.1
|
NPIPB8
|
nuclear pore complex interacting protein family member B8 |
| chr12_-_119804472 | 0.09 |
ENST00000678087.1
ENST00000677993.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr4_+_158521937 | 0.09 |
ENST00000343542.9
ENST00000470033.2 |
RXFP1
|
relaxin family peptide receptor 1 |
| chr17_-_51046868 | 0.09 |
ENST00000510283.5
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr12_+_26195313 | 0.09 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr2_+_172085499 | 0.09 |
ENST00000361725.5
ENST00000341900.6 |
DLX1
|
distal-less homeobox 1 |
| chr16_-_69384747 | 0.09 |
ENST00000566257.5
|
TERF2
|
telomeric repeat binding factor 2 |
| chr5_+_141408032 | 0.09 |
ENST00000520790.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr17_-_41060109 | 0.09 |
ENST00000391418.3
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chrX_-_120629936 | 0.09 |
ENST00000371313.2
ENST00000304661.6 |
C1GALT1C1
|
C1GALT1 specific chaperone 1 |
| chr6_+_39792298 | 0.09 |
ENST00000633794.1
ENST00000274867.9 |
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr19_-_51082883 | 0.09 |
ENST00000650543.2
|
KLK14
|
kallikrein related peptidase 14 |
| chr3_-_136752361 | 0.09 |
ENST00000480733.1
ENST00000629124.2 ENST00000383202.7 ENST00000236698.9 ENST00000434713.6 |
STAG1
|
stromal antigen 1 |
| chr15_-_34969688 | 0.08 |
ENST00000156471.10
|
AQR
|
aquarius intron-binding spliceosomal factor |
| chr11_+_121129964 | 0.08 |
ENST00000645008.1
|
TECTA
|
tectorin alpha |
| chr2_-_55296361 | 0.08 |
ENST00000647547.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr12_+_8157034 | 0.08 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A |
| chr8_-_11201799 | 0.08 |
ENST00000416569.3
|
XKR6
|
XK related 6 |
| chr14_-_24429665 | 0.08 |
ENST00000267406.11
|
CBLN3
|
cerebellin 3 precursor |
| chr3_+_46354072 | 0.08 |
ENST00000445132.3
ENST00000421659.1 |
CCR2
|
C-C motif chemokine receptor 2 |
| chrX_-_43973382 | 0.08 |
ENST00000642620.1
ENST00000647044.1 |
NDP
|
norrin cystine knot growth factor NDP |
| chr1_-_182400660 | 0.08 |
ENST00000367565.2
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr3_-_71304741 | 0.08 |
ENST00000484350.5
|
FOXP1
|
forkhead box P1 |
| chrX_-_19887585 | 0.08 |
ENST00000397821.8
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr5_+_50666660 | 0.08 |
ENST00000515175.6
|
PARP8
|
poly(ADP-ribose) polymerase family member 8 |
| chr15_-_32455634 | 0.08 |
ENST00000509311.7
|
GOLGA8O
|
golgin A8 family member O |
| chr3_+_152299570 | 0.08 |
ENST00000485910.5
ENST00000463374.5 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr7_-_6706843 | 0.08 |
ENST00000394917.3
ENST00000342651.9 ENST00000405858.6 |
ZNF12
|
zinc finger protein 12 |
| chr14_-_35121950 | 0.08 |
ENST00000554361.5
ENST00000261475.10 |
PPP2R3C
|
protein phosphatase 2 regulatory subunit B''gamma |
| chr1_+_171512032 | 0.08 |
ENST00000426496.6
|
PRRC2C
|
proline rich coiled-coil 2C |
| chr3_-_65597886 | 0.08 |
ENST00000460329.6
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_+_11806096 | 0.08 |
ENST00000312413.10
ENST00000346436.11 |
CLCN6
|
chloride voltage-gated channel 6 |
| chr1_+_36084079 | 0.08 |
ENST00000207457.8
|
TEKT2
|
tektin 2 |
| chrX_+_103356446 | 0.08 |
ENST00000372656.5
ENST00000372661.6 ENST00000646896.1 |
TCEAL9
|
transcription elongation factor A like 9 |
| chr17_-_37609361 | 0.08 |
ENST00000614941.4
ENST00000619541.4 ENST00000622045.4 ENST00000616179.4 ENST00000621136.4 ENST00000612223.5 ENST00000620424.1 |
SYNRG
|
synergin gamma |
| chr3_-_183099508 | 0.07 |
ENST00000476176.5
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 |
| chr1_-_120051714 | 0.07 |
ENST00000579475.7
|
NOTCH2
|
notch receptor 2 |
| chr3_-_179071742 | 0.07 |
ENST00000311417.7
ENST00000652290.1 |
ZMAT3
|
zinc finger matrin-type 3 |
| chr12_+_6772512 | 0.07 |
ENST00000441671.6
ENST00000203629.3 |
LAG3
|
lymphocyte activating 3 |
| chr1_-_169630115 | 0.07 |
ENST00000263686.11
ENST00000367788.6 |
SELP
|
selectin P |
| chr3_-_132684685 | 0.07 |
ENST00000512094.5
ENST00000632629.1 |
NPHP3
NPHP3-ACAD11
|
nephrocystin 3 NPHP3-ACAD11 readthrough (NMD candidate) |
| chrX_+_134807217 | 0.07 |
ENST00000482240.5
ENST00000475361.1 ENST00000645433.2 ENST00000494709.1 |
PABIR3
|
PABIR family member 3 |
| chr9_-_14314519 | 0.07 |
ENST00000397581.6
|
NFIB
|
nuclear factor I B |
| chr19_+_10086787 | 0.07 |
ENST00000590378.5
ENST00000397881.7 |
SHFL
|
shiftless antiviral inhibitor of ribosomal frameshifting |
| chr1_+_168576595 | 0.07 |
ENST00000367818.4
|
XCL1
|
X-C motif chemokine ligand 1 |
| chr16_+_22513523 | 0.07 |
ENST00000538606.5
ENST00000451409.5 ENST00000424340.5 ENST00000517539.5 ENST00000528249.5 |
NPIPB5
|
nuclear pore complex interacting protein family member B5 |
| chr16_-_21857418 | 0.07 |
ENST00000415645.6
|
NPIPB4
|
nuclear pore complex interacting protein family member B4 |
| chr15_+_32593456 | 0.07 |
ENST00000448387.6
ENST00000569659.5 |
GOLGA8N
|
golgin A8 family member N |
| chr3_-_158672612 | 0.07 |
ENST00000264265.4
|
LXN
|
latexin |
| chrX_-_13817027 | 0.06 |
ENST00000493677.5
ENST00000355135.6 ENST00000316715.9 |
GPM6B
|
glycoprotein M6B |
| chr12_-_58920465 | 0.06 |
ENST00000320743.8
|
LRIG3
|
leucine rich repeats and immunoglobulin like domains 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 0.6 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.2 | 1.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 0.7 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 2.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.4 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.3 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 0.6 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.1 | 1.0 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.8 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.3 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 0.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0048058 | compound eye corneal lens development(GO:0048058) |
| 0.0 | 0.3 | GO:2000768 | positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.1 | GO:0039023 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0021893 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.1 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0090264 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.1 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:2000537 | positive regulation of thymocyte migration(GO:2000412) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0061443 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell differentiation(GO:0061443) endocardial cushion cell fate commitment(GO:0061445) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:2000724 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0044557 | relaxation of smooth muscle(GO:0044557) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.3 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.4 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 2.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.2 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 1.4 | GO:0044291 | cell-cell contact zone(GO:0044291) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 1.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 5.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.0 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.0 | 0.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0043273 | CTPase activity(GO:0043273) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.0 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 1.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0033265 | choline binding(GO:0033265) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 4.5 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |