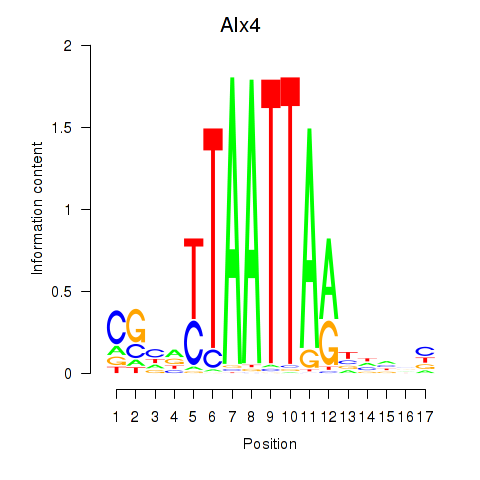
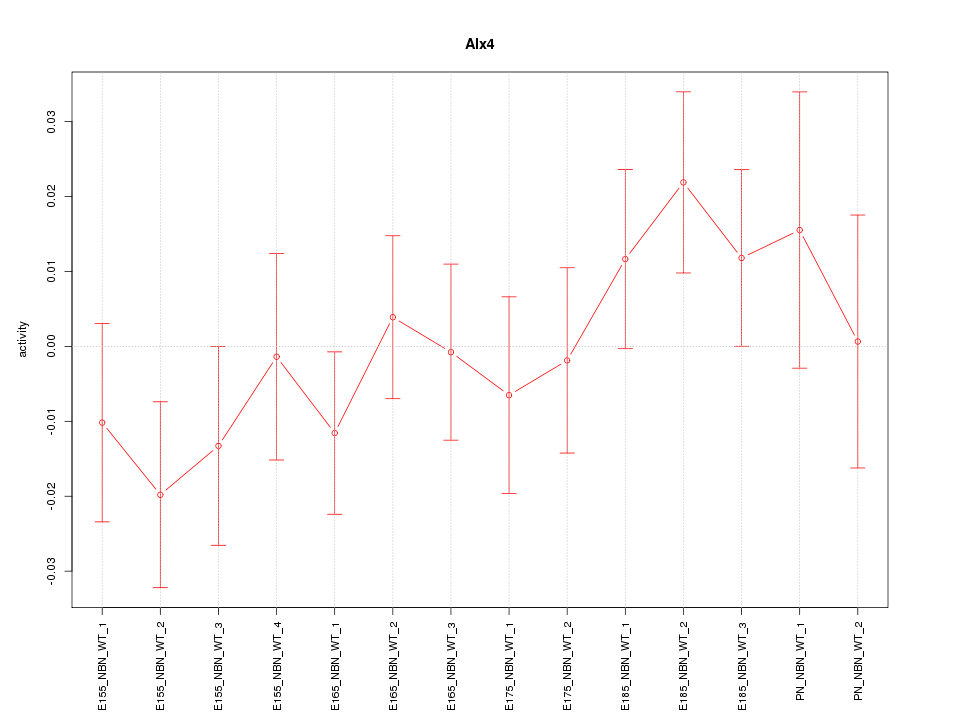
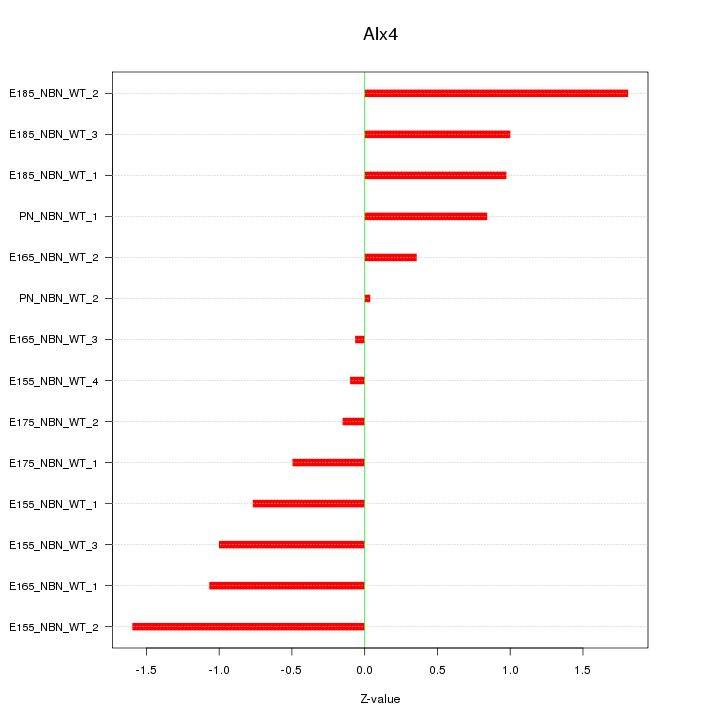
| 0.6 |
2.3 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.5 |
2.0 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.5 |
1.4 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.4 |
2.4 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.4 |
1.1 |
GO:1902498 |
regulation of protein autoubiquitination(GO:1902498) |
| 0.3 |
2.2 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.3 |
3.0 |
GO:0051923 |
sulfation(GO:0051923) |
| 0.3 |
0.8 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.2 |
0.7 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 |
1.9 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 |
0.6 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.2 |
0.5 |
GO:0072204 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.2 |
0.5 |
GO:0002538 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) olefin metabolic process(GO:1900673) |
| 0.2 |
0.5 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.2 |
0.5 |
GO:2000564 |
CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.2 |
0.8 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 |
0.9 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
1.9 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 |
0.5 |
GO:0043987 |
histone H3-S10 phosphorylation(GO:0043987) |
| 0.1 |
0.5 |
GO:0010749 |
regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.1 |
0.8 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.1 |
0.5 |
GO:1900170 |
prolactin signaling pathway(GO:0038161) negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 |
0.3 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 |
0.4 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 |
0.1 |
GO:1905050 |
positive regulation of metallopeptidase activity(GO:1905050) |
| 0.1 |
0.2 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 |
0.5 |
GO:1902255 |
positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.1 |
1.3 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.1 |
0.6 |
GO:1901162 |
serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 |
1.1 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
0.5 |
GO:0043504 |
mitochondrial DNA repair(GO:0043504) |
| 0.1 |
0.5 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 |
0.4 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.1 |
0.6 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
0.3 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.1 |
0.4 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.4 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.1 |
0.7 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
0.4 |
GO:0021555 |
midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 |
0.7 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
0.1 |
GO:1904956 |
regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 |
0.7 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 |
0.3 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.1 |
0.2 |
GO:0036518 |
chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 |
0.2 |
GO:0050787 |
glycolate metabolic process(GO:0009441) enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) positive regulation of superoxide dismutase activity(GO:1901671) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) glyoxal catabolic process(GO:1903190) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of removal of superoxide radicals(GO:1904833) regulation of cellular amino acid biosynthetic process(GO:2000282) positive regulation of cellular amino acid biosynthetic process(GO:2000284) positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 |
0.5 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 |
0.2 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 |
0.6 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 |
0.4 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.1 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 |
0.6 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.3 |
GO:1990086 |
lens fiber cell apoptotic process(GO:1990086) |
| 0.0 |
0.1 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.0 |
0.1 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 |
1.1 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.1 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.0 |
0.2 |
GO:0021856 |
cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.0 |
2.0 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
2.3 |
GO:0030500 |
regulation of bone mineralization(GO:0030500) |
| 0.0 |
0.4 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.0 |
0.3 |
GO:0006183 |
GTP biosynthetic process(GO:0006183) |
| 0.0 |
0.7 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
0.3 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.9 |
GO:0071385 |
cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 |
0.8 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |
| 0.0 |
0.1 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 |
2.3 |
GO:0019882 |
antigen processing and presentation(GO:0019882) |
| 0.0 |
0.2 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 |
0.1 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 |
1.0 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 |
0.2 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 |
0.6 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.2 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.2 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.3 |
GO:0006221 |
pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 |
0.3 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.4 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.0 |
0.3 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.0 |
0.1 |
GO:0052695 |
cellular glucuronidation(GO:0052695) |
| 0.0 |
0.1 |
GO:1901727 |
positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 |
0.1 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.8 |
GO:0016126 |
sterol biosynthetic process(GO:0016126) |
| 0.0 |
0.2 |
GO:0044406 |
adhesion of symbiont to host(GO:0044406) |
| 0.0 |
0.2 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 |
0.9 |
GO:0071774 |
response to fibroblast growth factor(GO:0071774) |
| 0.0 |
0.2 |
GO:1903427 |
negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 |
0.2 |
GO:0030497 |
fatty acid elongation(GO:0030497) |
| 0.0 |
0.4 |
GO:0010972 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 |
0.1 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.2 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |