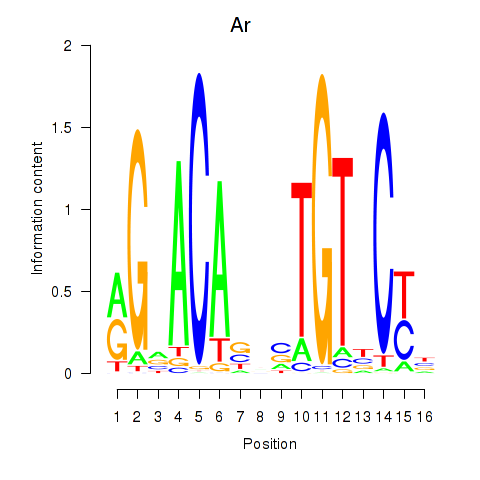
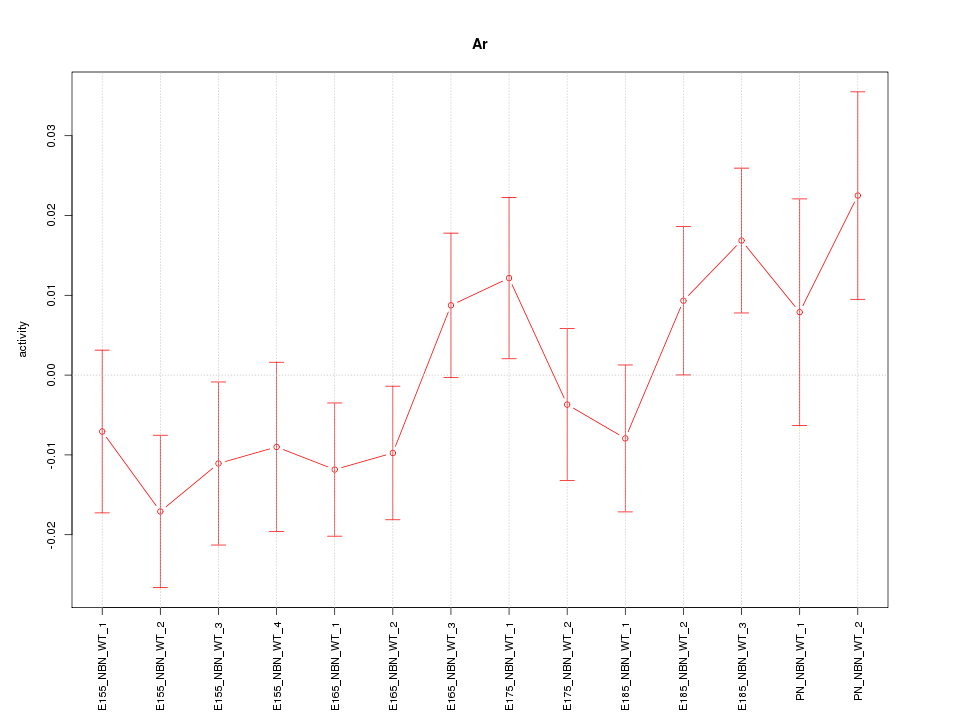
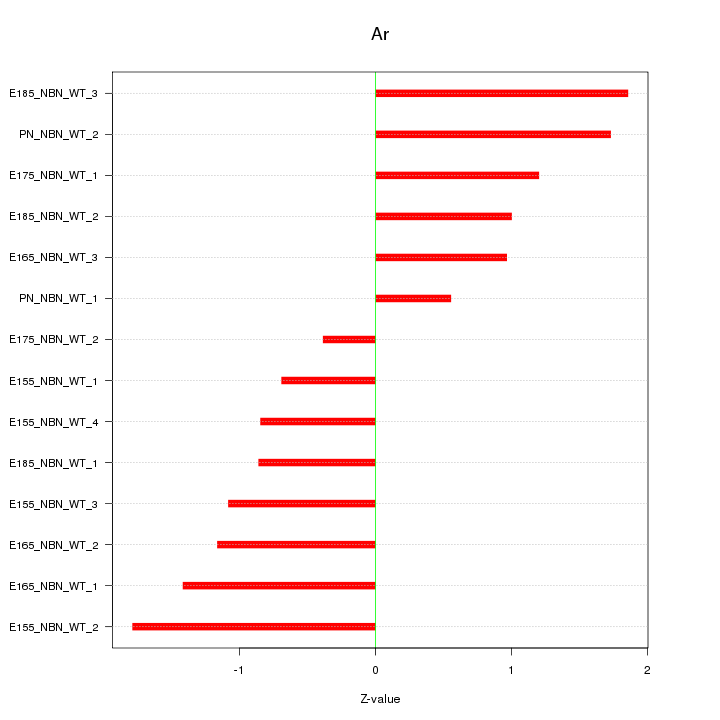
| 0.6 |
1.8 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.6 |
1.7 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.5 |
3.6 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.5 |
1.5 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.5 |
3.4 |
GO:0010273 |
detoxification of copper ion(GO:0010273) cellular response to zinc ion(GO:0071294) stress response to copper ion(GO:1990169) |
| 0.5 |
3.3 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.4 |
1.5 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.4 |
1.9 |
GO:0071672 |
negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.4 |
1.1 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.3 |
0.8 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 |
1.8 |
GO:0070253 |
somatostatin secretion(GO:0070253) |
| 0.2 |
1.5 |
GO:0071499 |
response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.2 |
0.7 |
GO:0021837 |
motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.2 |
1.7 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 |
3.8 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.2 |
2.0 |
GO:0051923 |
sulfation(GO:0051923) |
| 0.2 |
0.5 |
GO:2000170 |
positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 |
1.6 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 |
0.5 |
GO:0042560 |
10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 |
0.8 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.1 |
0.6 |
GO:0006547 |
histidine metabolic process(GO:0006547) |
| 0.1 |
0.6 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.1 |
0.4 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.1 |
0.2 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 |
0.6 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 0.1 |
0.6 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 |
0.6 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
0.4 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 |
1.9 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.1 |
0.5 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 |
0.3 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.1 |
0.4 |
GO:0071449 |
cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 |
0.2 |
GO:1902445 |
regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 |
0.4 |
GO:0051182 |
coenzyme transport(GO:0051182) |
| 0.1 |
1.5 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.1 |
0.4 |
GO:0010745 |
negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 |
0.8 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.1 |
0.2 |
GO:1904017 |
response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 |
0.1 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 |
0.2 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 0.1 |
0.2 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 |
0.7 |
GO:0036297 |
interstrand cross-link repair(GO:0036297) |
| 0.1 |
0.2 |
GO:2001106 |
regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 |
0.1 |
GO:2000157 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 |
0.1 |
GO:0035811 |
negative regulation of urine volume(GO:0035811) |
| 0.1 |
1.4 |
GO:0010866 |
regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 |
0.4 |
GO:0070475 |
rRNA base methylation(GO:0070475) |
| 0.0 |
0.5 |
GO:0006490 |
oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 |
5.4 |
GO:0016052 |
carbohydrate catabolic process(GO:0016052) |
| 0.0 |
0.5 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.0 |
0.4 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.0 |
0.2 |
GO:0098789 |
pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 |
0.2 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.0 |
0.2 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 |
0.4 |
GO:2001135 |
regulation of endocytic recycling(GO:2001135) |
| 0.0 |
0.6 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.1 |
GO:0019441 |
tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) quinolinate biosynthetic process(GO:0019805) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.0 |
1.0 |
GO:0051497 |
negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 |
0.4 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.9 |
GO:0035458 |
cellular response to interferon-beta(GO:0035458) |
| 0.0 |
0.1 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.0 |
0.2 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.7 |
GO:0006346 |
methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 |
0.3 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 |
1.7 |
GO:0048747 |
muscle fiber development(GO:0048747) |
| 0.0 |
0.1 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.0 |
0.1 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.0 |
0.4 |
GO:0072520 |
seminiferous tubule development(GO:0072520) |
| 0.0 |
0.8 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.0 |
0.1 |
GO:0016102 |
retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 |
0.5 |
GO:0006760 |
folic acid-containing compound metabolic process(GO:0006760) |
| 0.0 |
0.2 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
1.1 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
0.1 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 |
0.1 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 |
0.5 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.0 |
0.2 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.0 |
0.1 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.5 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.2 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.3 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
2.3 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.5 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.2 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.5 |
GO:0070303 |
negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 |
0.1 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.0 |
0.4 |
GO:0043303 |
mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 |
0.1 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.1 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |