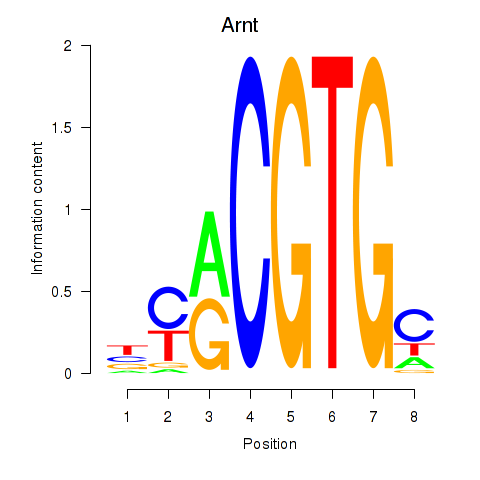
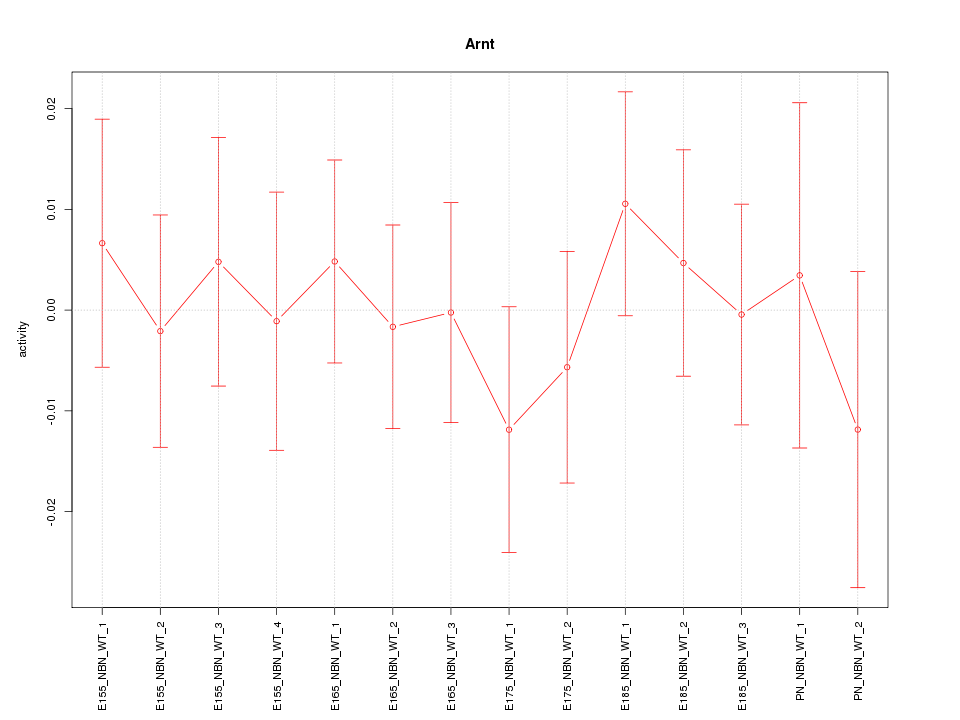
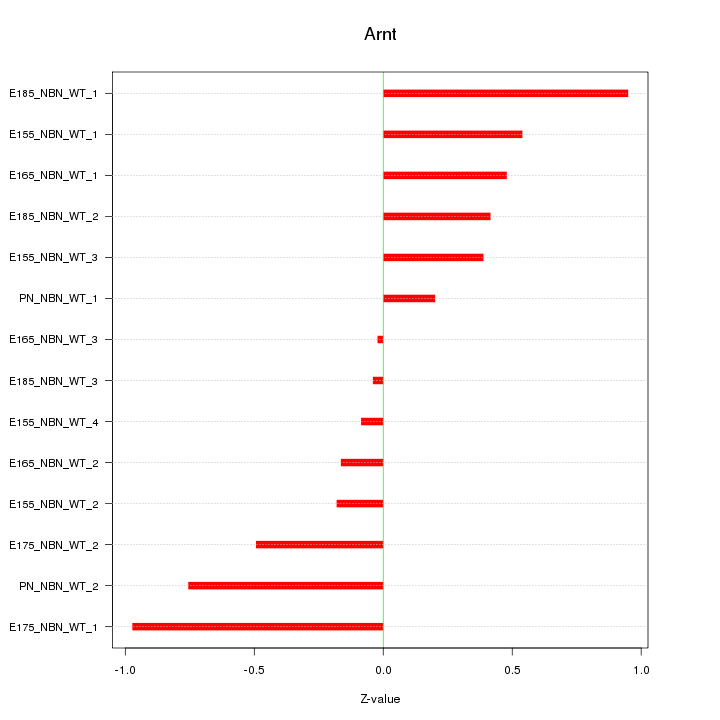
| 0.1 |
0.3 |
GO:0048389 |
BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) intermediate mesoderm development(GO:0048389) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) pattern specification involved in mesonephros development(GO:0061227) regulation of hepatocyte differentiation(GO:0070366) comma-shaped body morphogenesis(GO:0072049) S-shaped body morphogenesis(GO:0072050) anterior/posterior pattern specification involved in kidney development(GO:0072098) negative regulation of glomerular mesangial cell proliferation(GO:0072125) ureter urothelium development(GO:0072190) negative regulation of glomerulus development(GO:0090194) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 |
0.3 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 |
0.1 |
GO:0044830 |
modulation by host of viral RNA genome replication(GO:0044830) |
| 0.1 |
0.3 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of cellular pH reduction(GO:0032847) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 |
0.9 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
0.2 |
GO:0045168 |
cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.1 |
0.2 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 |
0.3 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.1 |
0.2 |
GO:0070428 |
granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 |
0.2 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 |
0.2 |
GO:0019405 |
hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) fructose biosynthetic process(GO:0046370) |
| 0.1 |
0.2 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 |
0.2 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 |
0.1 |
GO:0010248 |
B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 |
0.2 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.0 |
0.2 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 |
0.3 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 |
0.2 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 |
0.1 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.0 |
0.0 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.0 |
0.2 |
GO:0030214 |
hyaluronan catabolic process(GO:0030214) |
| 0.0 |
0.1 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.0 |
0.1 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.0 |
0.4 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.1 |
GO:0034144 |
negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 |
0.5 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 |
0.1 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 |
0.1 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 |
0.2 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.0 |
0.2 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 |
0.2 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 |
0.1 |
GO:0098908 |
regulation of neuronal action potential(GO:0098908) |
| 0.0 |
0.1 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.1 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.1 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.1 |
GO:0051004 |
regulation of lipoprotein lipase activity(GO:0051004) |
| 0.0 |
0.1 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 |
0.2 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.4 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
0.1 |
GO:0010917 |
negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 |
0.1 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) |
| 0.0 |
0.3 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.2 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.1 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 |
0.1 |
GO:0040031 |
snRNA modification(GO:0040031) |
| 0.0 |
0.1 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.0 |
0.1 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.2 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.3 |
GO:0097284 |
hepatocyte apoptotic process(GO:0097284) |
| 0.0 |
0.0 |
GO:0046398 |
UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 |
0.1 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.6 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.0 |
0.1 |
GO:0015744 |
succinate transport(GO:0015744) |
| 0.0 |
0.2 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.1 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 |
0.1 |
GO:0002922 |
positive regulation of humoral immune response(GO:0002922) progesterone receptor signaling pathway(GO:0050847) |
| 0.0 |
0.1 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.0 |
0.1 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.0 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 |
0.1 |
GO:0045053 |
protein retention in Golgi apparatus(GO:0045053) |
| 0.0 |
0.0 |
GO:0046084 |
adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 |
0.1 |
GO:0044829 |
negative regulation by host of viral genome replication(GO:0044828) positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 |
0.0 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.0 |
0.1 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.4 |
GO:0035116 |
embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 |
0.1 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |