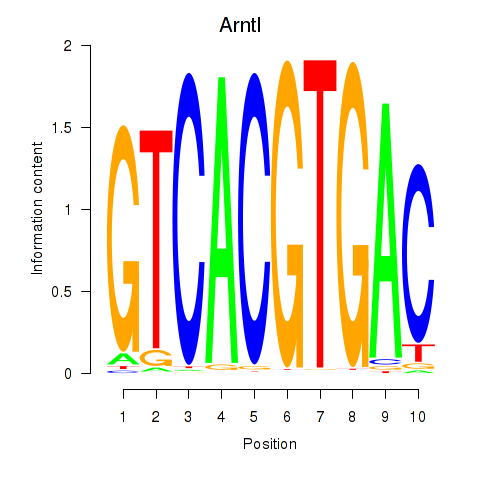
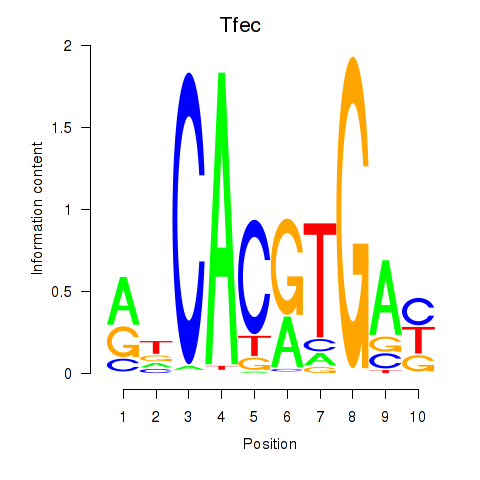
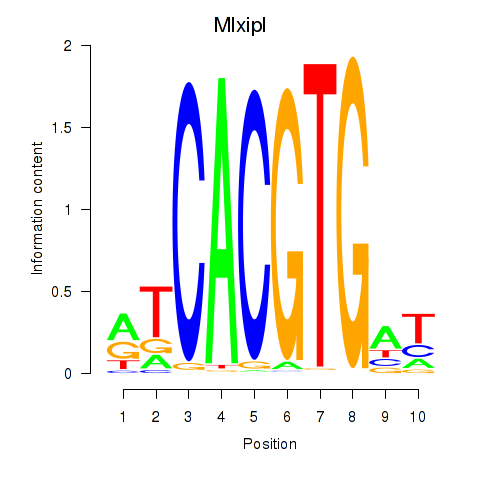
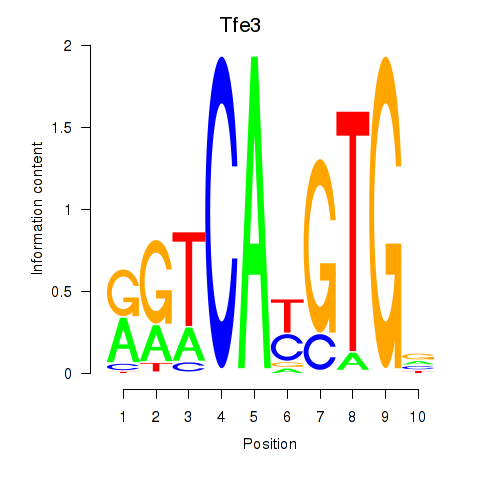
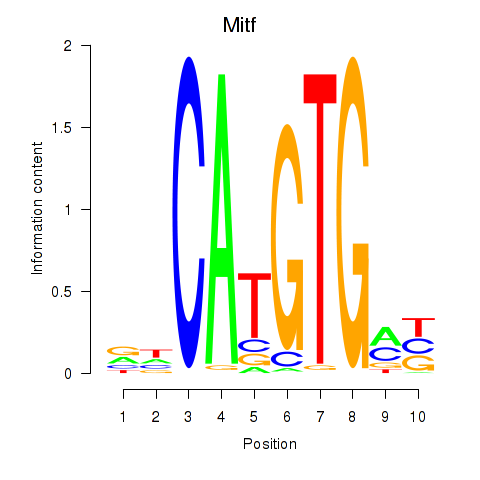
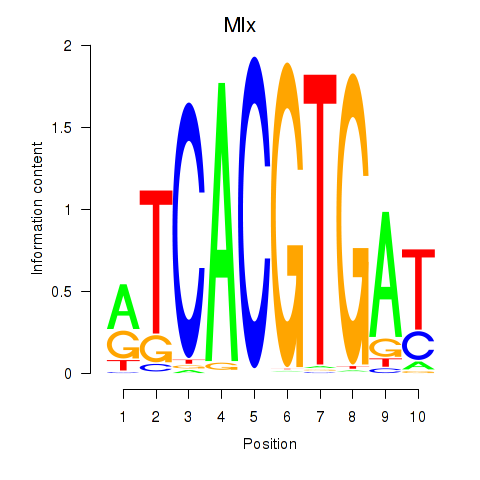
| 0.7 |
2.0 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.5 |
1.6 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.5 |
1.4 |
GO:1904706 |
negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.5 |
2.3 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.4 |
1.7 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.4 |
1.3 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.3 |
0.9 |
GO:0010248 |
B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.3 |
0.9 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.3 |
1.2 |
GO:1900426 |
positive regulation of defense response to bacterium(GO:1900426) |
| 0.3 |
0.8 |
GO:0006001 |
fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.3 |
1.5 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.2 |
0.9 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.2 |
0.7 |
GO:0071873 |
response to norepinephrine(GO:0071873) |
| 0.2 |
0.7 |
GO:0043181 |
vacuolar sequestering(GO:0043181) |
| 0.2 |
1.1 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 |
1.5 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 |
0.6 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 |
1.6 |
GO:0071907 |
determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.2 |
0.6 |
GO:0030421 |
defecation(GO:0030421) |
| 0.2 |
0.6 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.2 |
0.4 |
GO:0048597 |
post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.2 |
4.8 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 |
0.5 |
GO:0009177 |
deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.2 |
0.5 |
GO:0046032 |
ADP catabolic process(GO:0046032) |
| 0.2 |
1.1 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 |
2.8 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 |
0.7 |
GO:2001204 |
regulation of osteoclast development(GO:2001204) |
| 0.1 |
0.4 |
GO:0006285 |
base-excision repair, AP site formation(GO:0006285) |
| 0.1 |
0.4 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 |
0.4 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.1 |
0.9 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.1 |
0.5 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 |
0.4 |
GO:0070476 |
rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 |
0.9 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.1 |
2.4 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.1 |
0.4 |
GO:0070625 |
zymogen granule exocytosis(GO:0070625) |
| 0.1 |
0.4 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 |
0.6 |
GO:0060620 |
regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.1 |
0.4 |
GO:0060785 |
regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 |
1.2 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 |
0.8 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.1 |
0.5 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
1.6 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 |
0.5 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.1 |
0.6 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.1 |
0.3 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.1 |
0.5 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.1 |
0.5 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 |
0.6 |
GO:0045627 |
positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.1 |
0.3 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.1 |
0.3 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 |
0.6 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.1 |
0.3 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) |
| 0.1 |
0.4 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 |
0.2 |
GO:0010046 |
response to mycotoxin(GO:0010046) |
| 0.1 |
0.3 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.1 |
0.4 |
GO:0034184 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 |
0.3 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 |
1.1 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 0.1 |
0.4 |
GO:2001245 |
regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 |
0.1 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.1 |
0.3 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 |
0.4 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.1 |
0.4 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.1 |
0.3 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 0.1 |
0.3 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 |
0.2 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.1 |
0.5 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 |
0.7 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
0.2 |
GO:0009193 |
pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 |
0.5 |
GO:0010917 |
negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.1 |
0.3 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.1 |
0.4 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 |
0.2 |
GO:0010936 |
negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 |
0.1 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 |
0.6 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.1 |
0.1 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.1 |
0.1 |
GO:0090080 |
positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 |
0.1 |
GO:0001788 |
antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.1 |
0.4 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.1 |
0.3 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 |
0.9 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.1 |
2.6 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.1 |
0.3 |
GO:0007042 |
lysosomal lumen acidification(GO:0007042) |
| 0.1 |
0.3 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.1 |
0.5 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.1 |
0.8 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.1 |
0.3 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 |
0.1 |
GO:0051342 |
regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 |
0.3 |
GO:0002924 |
negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 |
0.9 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.1 |
0.1 |
GO:0032817 |
regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.1 |
0.4 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 |
0.5 |
GO:0010944 |
negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 |
0.7 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.1 |
0.2 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.1 |
0.3 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 |
0.2 |
GO:0046121 |
deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 |
0.2 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.1 |
0.4 |
GO:2000781 |
positive regulation of double-strand break repair(GO:2000781) |
| 0.1 |
0.6 |
GO:0051533 |
positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 |
0.2 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.1 |
0.2 |
GO:0071105 |
negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.1 |
0.2 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 |
1.4 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 |
0.8 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.1 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 |
0.1 |
GO:1902498 |
regulation of protein autoubiquitination(GO:1902498) |
| 0.1 |
2.1 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.1 |
0.2 |
GO:1904569 |
regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 |
0.7 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.1 |
0.2 |
GO:0009181 |
purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 |
0.6 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.1 |
0.4 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 |
0.4 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.1 |
0.3 |
GO:2000394 |
positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 |
0.1 |
GO:0019043 |
establishment of viral latency(GO:0019043) |
| 0.0 |
0.1 |
GO:0060066 |
oviduct development(GO:0060066) |
| 0.0 |
0.6 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.0 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 |
0.1 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.0 |
0.4 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.0 |
0.8 |
GO:0051974 |
negative regulation of telomerase activity(GO:0051974) |
| 0.0 |
0.3 |
GO:0046502 |
uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 |
0.2 |
GO:0015744 |
succinate transport(GO:0015744) |
| 0.0 |
0.2 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 |
0.2 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 |
0.2 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 |
0.2 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.3 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.2 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 |
0.0 |
GO:2000382 |
positive regulation of mesoderm development(GO:2000382) |
| 0.0 |
0.4 |
GO:0006590 |
thyroid hormone generation(GO:0006590) |
| 0.0 |
0.8 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.3 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.0 |
0.3 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.0 |
0.6 |
GO:0097154 |
GABAergic neuron differentiation(GO:0097154) |
| 0.0 |
0.6 |
GO:0030011 |
maintenance of cell polarity(GO:0030011) |
| 0.0 |
0.2 |
GO:0040031 |
snRNA modification(GO:0040031) |
| 0.0 |
0.2 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.1 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 |
0.1 |
GO:0040009 |
regulation of growth rate(GO:0040009) |
| 0.0 |
0.1 |
GO:0000965 |
mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 |
0.1 |
GO:2000170 |
positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 |
0.0 |
GO:0000338 |
protein deneddylation(GO:0000338) |
| 0.0 |
0.4 |
GO:2000653 |
regulation of genetic imprinting(GO:2000653) |
| 0.0 |
1.0 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.7 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.2 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.0 |
0.4 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.0 |
0.2 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
1.1 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.2 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.0 |
0.1 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.0 |
0.1 |
GO:0043987 |
histone H3-S10 phosphorylation(GO:0043987) histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 |
0.1 |
GO:1902044 |
regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.0 |
0.3 |
GO:1901387 |
positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 |
0.1 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.0 |
0.5 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.3 |
GO:2000467 |
positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 |
1.2 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.0 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.5 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 |
0.1 |
GO:1904417 |
regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 |
0.4 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 |
0.4 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.1 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.0 |
0.1 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 |
0.1 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.0 |
0.1 |
GO:0032211 |
negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 |
0.2 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.0 |
0.1 |
GO:0061724 |
lipophagy(GO:0061724) |
| 0.0 |
0.5 |
GO:0010666 |
positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 |
0.1 |
GO:0070942 |
neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) |
| 0.0 |
0.2 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 |
0.3 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
0.1 |
GO:1902512 |
positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 |
0.1 |
GO:0015870 |
acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.0 |
0.0 |
GO:0060160 |
negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 |
0.9 |
GO:0010866 |
regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 |
1.4 |
GO:0048255 |
mRNA stabilization(GO:0048255) |
| 0.0 |
0.2 |
GO:0032471 |
negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.0 |
0.4 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.2 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 |
0.1 |
GO:0045776 |
negative regulation of blood pressure(GO:0045776) |
| 0.0 |
0.1 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.0 |
0.1 |
GO:0035927 |
RNA import into mitochondrion(GO:0035927) |
| 0.0 |
0.2 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.0 |
0.8 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.2 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.5 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.0 |
0.1 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.0 |
0.1 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.0 |
0.1 |
GO:1903774 |
positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 |
0.3 |
GO:0006833 |
water transport(GO:0006833) |
| 0.0 |
0.1 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 |
0.1 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 |
0.1 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 |
0.1 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.0 |
0.2 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 |
0.2 |
GO:0070307 |
lens fiber cell development(GO:0070307) |
| 0.0 |
0.1 |
GO:2000525 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 |
0.9 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:0046645 |
positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 |
0.1 |
GO:0006545 |
glycine biosynthetic process(GO:0006545) |
| 0.0 |
0.2 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.0 |
0.0 |
GO:0060754 |
positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 |
0.0 |
GO:0010873 |
positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 |
0.2 |
GO:0051532 |
regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 |
0.1 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.0 |
0.1 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.0 |
0.3 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.5 |
GO:0060441 |
epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 |
0.2 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 |
0.1 |
GO:0043096 |
purine nucleobase salvage(GO:0043096) |
| 0.0 |
0.1 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.1 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.8 |
GO:0048011 |
neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 |
0.1 |
GO:0071475 |
cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 |
0.1 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.1 |
GO:0046643 |
regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 |
0.1 |
GO:0046294 |
formaldehyde catabolic process(GO:0046294) |
| 0.0 |
0.1 |
GO:0042219 |
cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 |
0.2 |
GO:1901970 |
positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.0 |
0.2 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 |
0.1 |
GO:0045471 |
response to ethanol(GO:0045471) |
| 0.0 |
0.1 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 |
0.1 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.0 |
0.0 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 |
0.2 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.0 |
0.3 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.7 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.1 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 |
0.0 |
GO:0043489 |
RNA stabilization(GO:0043489) |
| 0.0 |
0.1 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.0 |
0.2 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.0 |
GO:0035106 |
operant conditioning(GO:0035106) |
| 0.0 |
0.1 |
GO:1990046 |
positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 0.0 |
0.1 |
GO:0046013 |
T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 |
0.1 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 |
0.1 |
GO:0051204 |
protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 |
0.3 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 |
0.1 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 |
0.1 |
GO:0021586 |
pons maturation(GO:0021586) |
| 0.0 |
0.1 |
GO:0046654 |
tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 |
0.1 |
GO:0060346 |
bone trabecula formation(GO:0060346) |
| 0.0 |
0.2 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.1 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.0 |
0.1 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 |
0.0 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.0 |
GO:0034287 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 |
0.1 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.0 |
0.1 |
GO:0009988 |
cell-cell recognition(GO:0009988) |
| 0.0 |
0.2 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 |
0.1 |
GO:0052695 |
cellular glucuronidation(GO:0052695) |
| 0.0 |
0.1 |
GO:0034088 |
maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 |
1.7 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
0.1 |
GO:0090220 |
meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 |
0.2 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
0.1 |
GO:0042228 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) interleukin-8 biosynthetic process(GO:0042228) regulation of interleukin-8 biosynthetic process(GO:0045414) |
| 0.0 |
0.1 |
GO:0033313 |
meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 |
0.0 |
GO:0042726 |
flavin-containing compound metabolic process(GO:0042726) |
| 0.0 |
0.1 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
0.2 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.1 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.0 |
0.0 |
GO:0045629 |
negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 |
0.1 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.1 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 |
0.2 |
GO:0014046 |
dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 |
0.1 |
GO:0006982 |
response to lipid hydroperoxide(GO:0006982) |
| 0.0 |
0.0 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 |
0.1 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 |
0.0 |
GO:0010593 |
negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 |
0.2 |
GO:0097011 |
cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 |
0.4 |
GO:0009303 |
rRNA transcription(GO:0009303) |
| 0.0 |
0.2 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 |
0.1 |
GO:0000727 |
double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 |
0.1 |
GO:0038030 |
non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 |
0.1 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.0 |
0.1 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.0 |
GO:1901492 |
positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 |
0.1 |
GO:0006183 |
GTP biosynthetic process(GO:0006183) |
| 0.0 |
0.1 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 |
0.4 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.2 |
GO:0045669 |
positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 |
0.1 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.0 |
0.4 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.3 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.2 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 |
0.0 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 |
0.0 |
GO:0036518 |
chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 |
0.0 |
GO:0042373 |
vitamin K metabolic process(GO:0042373) |
| 0.0 |
0.7 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.3 |
GO:0001913 |
T cell mediated cytotoxicity(GO:0001913) |
| 0.0 |
0.1 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 |
0.1 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 |
0.1 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 |
0.1 |
GO:0019478 |
D-amino acid catabolic process(GO:0019478) |
| 0.0 |
0.1 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.0 |
GO:0071073 |
positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 |
0.0 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.1 |
GO:0006048 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 |
0.0 |
GO:0010920 |
negative regulation of inositol phosphate biosynthetic process(GO:0010920) testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 |
0.0 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.0 |
0.1 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 |
0.0 |
GO:0034144 |
negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 |
0.5 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.1 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.3 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.0 |
GO:0045006 |
DNA deamination(GO:0045006) |
| 0.0 |
0.1 |
GO:0035994 |
response to muscle stretch(GO:0035994) |
| 0.0 |
0.1 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.2 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
0.1 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.0 |
0.1 |
GO:1902414 |
protein localization to cell junction(GO:1902414) |
| 0.0 |
0.3 |
GO:0051495 |
positive regulation of cytoskeleton organization(GO:0051495) |
| 0.0 |
0.1 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.0 |
0.1 |
GO:1902224 |
cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.0 |
0.1 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 |
0.0 |
GO:2000767 |
positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 |
0.0 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 |
0.1 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.0 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 |
0.7 |
GO:0032088 |
negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 |
0.0 |
GO:0044332 |
Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 |
0.0 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.1 |
GO:0043383 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.2 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 |
0.3 |
GO:0045948 |
positive regulation of translational initiation(GO:0045948) |
| 0.0 |
0.1 |
GO:0033089 |
positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 |
0.1 |
GO:0030423 |
targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 |
0.1 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 |
0.1 |
GO:2000809 |
positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 |
0.3 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.1 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.0 |
0.0 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 |
0.1 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.0 |
GO:0048208 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 |
0.4 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.0 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.0 |
0.1 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 |
0.1 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 |
0.2 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.0 |
GO:0046544 |
development of secondary male sexual characteristics(GO:0046544) |
| 0.0 |
0.1 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 |
0.2 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.3 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.0 |
0.3 |
GO:1902750 |
negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.0 |
0.1 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.0 |
0.1 |
GO:0021554 |
optic nerve development(GO:0021554) |
| 0.0 |
0.0 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) |
| 0.0 |
0.0 |
GO:0032687 |
negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 |
0.1 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 |
0.0 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 |
0.2 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 |
0.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.3 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.3 |
GO:1903146 |
regulation of mitophagy(GO:1903146) |
| 0.0 |
0.1 |
GO:0003433 |
chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 |
0.6 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.1 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.0 |
GO:0071500 |
cellular response to nitrosative stress(GO:0071500) |
| 0.0 |
0.6 |
GO:0048663 |
neuron fate commitment(GO:0048663) |
| 0.0 |
0.0 |
GO:1901421 |
positive regulation of response to alcohol(GO:1901421) |
| 0.0 |
0.0 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 |
0.1 |
GO:0045839 |
negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 |
0.0 |
GO:0007096 |
regulation of exit from mitosis(GO:0007096) |
| 0.0 |
0.1 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 |
0.1 |
GO:0070203 |
regulation of establishment of protein localization to chromosome(GO:0070202) regulation of establishment of protein localization to telomere(GO:0070203) positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.0 |
0.1 |
GO:0010748 |
regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 |
0.1 |
GO:2001241 |
positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 |
0.1 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.1 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.1 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.0 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 |
0.0 |
GO:0015868 |
purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.0 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 |
0.0 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.1 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |