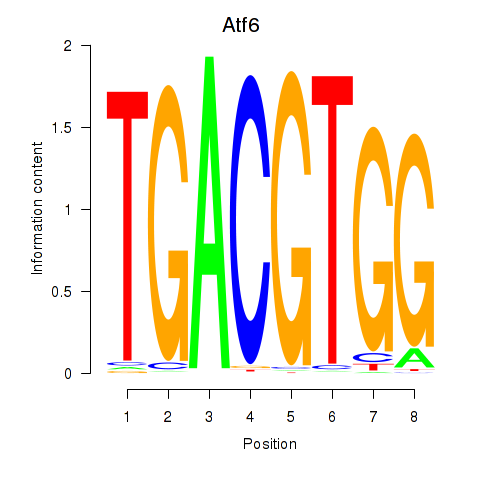
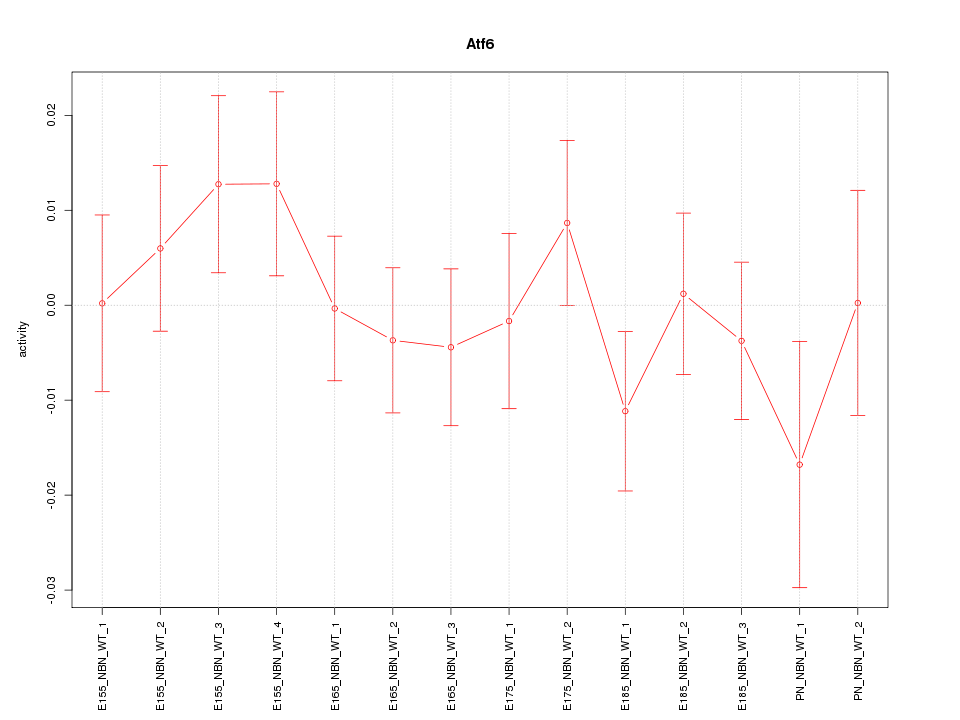
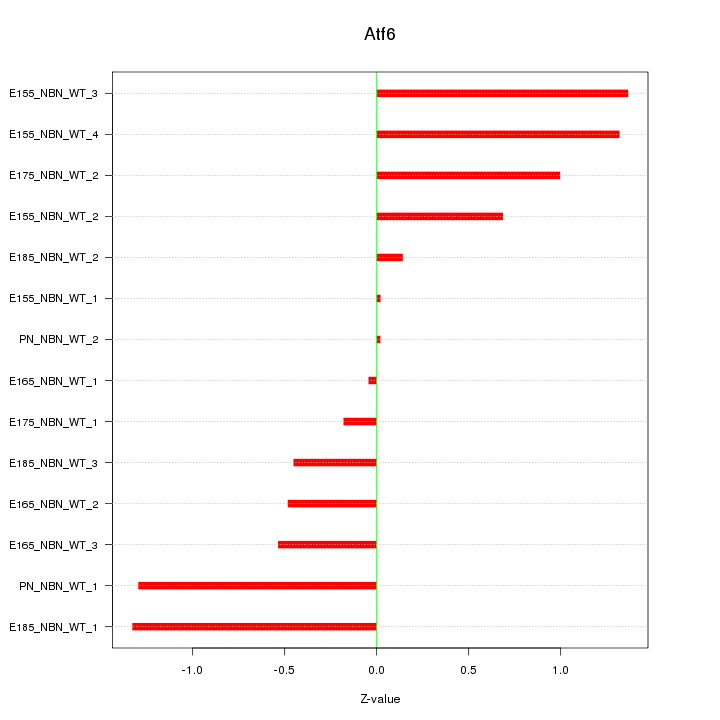
| 0.8 |
6.2 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.6 |
1.9 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 0.3 |
0.9 |
GO:0036353 |
histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.3 |
0.3 |
GO:0090298 |
negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.3 |
4.7 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 |
0.7 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 |
0.7 |
GO:0003195 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.2 |
1.1 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 |
1.6 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 |
1.2 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 |
0.5 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.1 |
1.0 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.1 |
0.4 |
GO:0043379 |
memory T cell differentiation(GO:0043379) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 |
0.4 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 |
0.4 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.1 |
0.5 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.1 |
0.4 |
GO:0071336 |
lung growth(GO:0060437) positive regulation of fat cell proliferation(GO:0070346) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.1 |
0.4 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.1 |
1.7 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
0.5 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.1 |
0.7 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 |
0.3 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.1 |
0.3 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 |
0.3 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.1 |
0.6 |
GO:0048133 |
NK T cell differentiation(GO:0001865) germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.1 |
0.7 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
0.4 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
0.2 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 |
0.3 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 |
0.3 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 |
1.4 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.1 |
0.6 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.1 |
0.5 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 |
2.2 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.1 |
1.4 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 |
0.3 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) |
| 0.1 |
0.6 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 |
0.2 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 |
0.2 |
GO:0060067 |
cervix development(GO:0060067) comma-shaped body morphogenesis(GO:0072049) S-shaped body morphogenesis(GO:0072050) anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 |
0.3 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 |
0.8 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 |
0.2 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 |
0.3 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 |
0.5 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 |
0.3 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.1 |
1.1 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
0.2 |
GO:0097709 |
columnar/cuboidal epithelial cell maturation(GO:0002069) connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.1 |
0.4 |
GO:0021937 |
cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 |
0.4 |
GO:0035067 |
negative regulation of histone acetylation(GO:0035067) |
| 0.0 |
0.1 |
GO:2000819 |
regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 |
0.1 |
GO:0032916 |
positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 |
0.1 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 |
0.7 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.0 |
0.4 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.7 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) |
| 0.0 |
1.1 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 |
0.2 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.1 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.4 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 |
0.2 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.0 |
0.1 |
GO:0070103 |
interleukin-4-mediated signaling pathway(GO:0035771) tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.0 |
0.1 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.0 |
0.1 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.0 |
0.2 |
GO:1904008 |
response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.0 |
0.1 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.0 |
0.1 |
GO:0006083 |
acetate metabolic process(GO:0006083) |
| 0.0 |
0.4 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 |
0.3 |
GO:0044351 |
macropinocytosis(GO:0044351) |
| 0.0 |
0.1 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.0 |
0.4 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.0 |
0.2 |
GO:0006982 |
response to lipid hydroperoxide(GO:0006982) |
| 0.0 |
0.6 |
GO:0007289 |
spermatid nucleus differentiation(GO:0007289) |
| 0.0 |
0.7 |
GO:0060416 |
response to growth hormone(GO:0060416) |
| 0.0 |
0.2 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.3 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 |
0.1 |
GO:1900147 |
Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 |
0.1 |
GO:0032474 |
otolith morphogenesis(GO:0032474) |
| 0.0 |
0.2 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.0 |
0.1 |
GO:0002904 |
positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 |
0.2 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.0 |
0.4 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.0 |
0.5 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.5 |
GO:0042438 |
manganese ion transport(GO:0006828) melanin biosynthetic process(GO:0042438) |
| 0.0 |
0.2 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 |
0.4 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.0 |
0.0 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 |
0.1 |
GO:0090091 |
regulation of microvillus assembly(GO:0032534) positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.1 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.1 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 |
0.2 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.2 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.1 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 |
0.7 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.3 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.4 |
GO:0048643 |
positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 |
0.1 |
GO:1900109 |
regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 |
0.3 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
0.4 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.1 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
0.7 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 |
0.1 |
GO:0002829 |
negative regulation of type 2 immune response(GO:0002829) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 |
0.1 |
GO:0045003 |
DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 |
0.2 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.0 |
0.0 |
GO:0071072 |
negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 |
0.1 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.1 |
GO:0006824 |
cobalt ion transport(GO:0006824) |
| 0.0 |
0.3 |
GO:0042059 |
negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 |
0.3 |
GO:0002063 |
chondrocyte development(GO:0002063) |
| 0.0 |
0.2 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.2 |
GO:0009191 |
ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 |
0.1 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 |
0.1 |
GO:0003338 |
metanephros morphogenesis(GO:0003338) |
| 0.0 |
0.1 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 |
0.1 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.0 |
GO:0055005 |
ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 |
0.1 |
GO:0002862 |
negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 |
0.1 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.0 |
0.5 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.1 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 |
0.7 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |