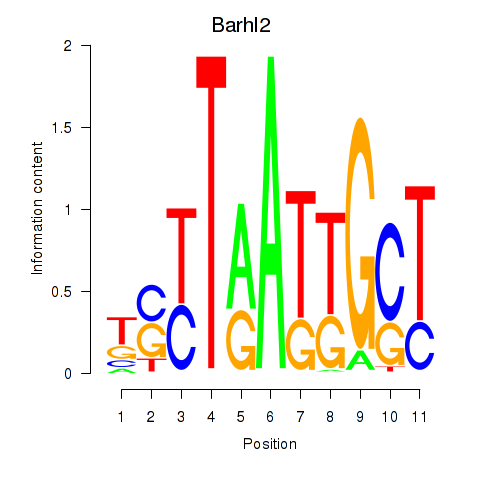
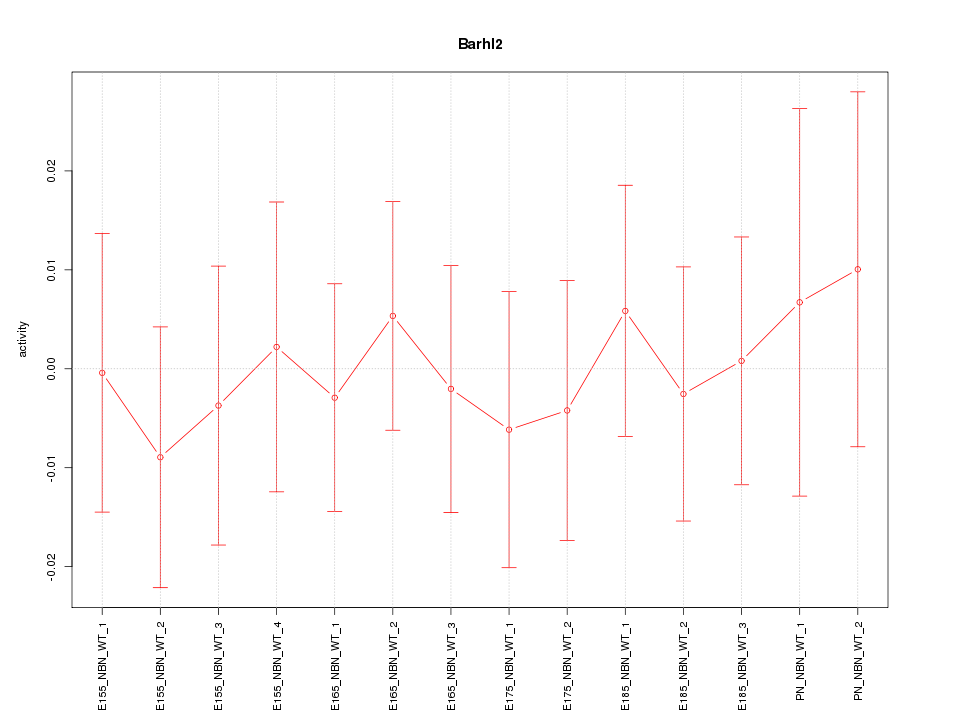
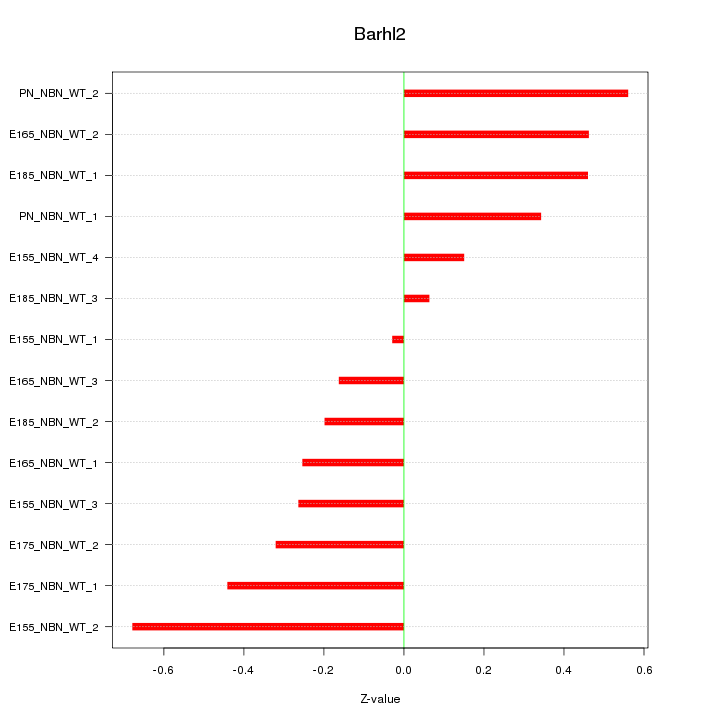
| 0.2 |
0.6 |
GO:0072223 |
metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.2 |
0.6 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.1 |
0.4 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.1 |
0.5 |
GO:1902990 |
mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.1 |
0.5 |
GO:1903463 |
mitotic cell cycle phase(GO:0098763) regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.1 |
0.4 |
GO:0035880 |
embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 |
0.3 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.1 |
0.3 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.1 |
0.4 |
GO:2000974 |
negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 |
0.3 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 |
0.5 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.1 |
0.2 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) renal vesicle induction(GO:0072034) |
| 0.1 |
0.6 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 |
0.3 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 |
0.7 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.5 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.7 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
0.1 |
GO:1903026 |
negative regulation of CREB transcription factor activity(GO:0032792) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 |
0.1 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 |
0.2 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 |
0.3 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.0 |
0.5 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.3 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.2 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.8 |
GO:0031114 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 |
0.1 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 |
0.1 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.0 |
0.1 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) germline stem cell asymmetric division(GO:0098728) |
| 0.0 |
0.1 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 |
0.2 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.0 |
0.1 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 |
0.2 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.3 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.2 |
GO:0042761 |
very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 |
0.2 |
GO:0032287 |
peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 |
0.1 |
GO:0030174 |
regulation of DNA-dependent DNA replication initiation(GO:0030174) mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 |
0.1 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.4 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |