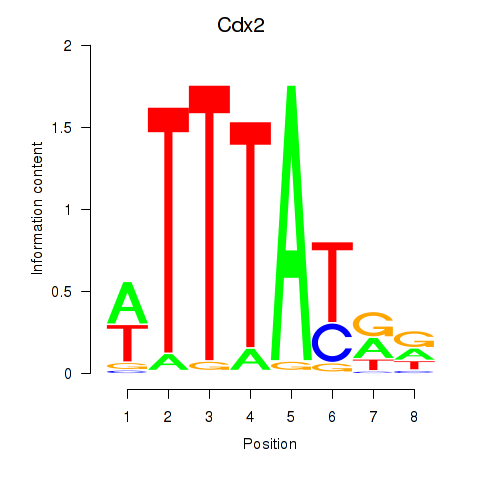
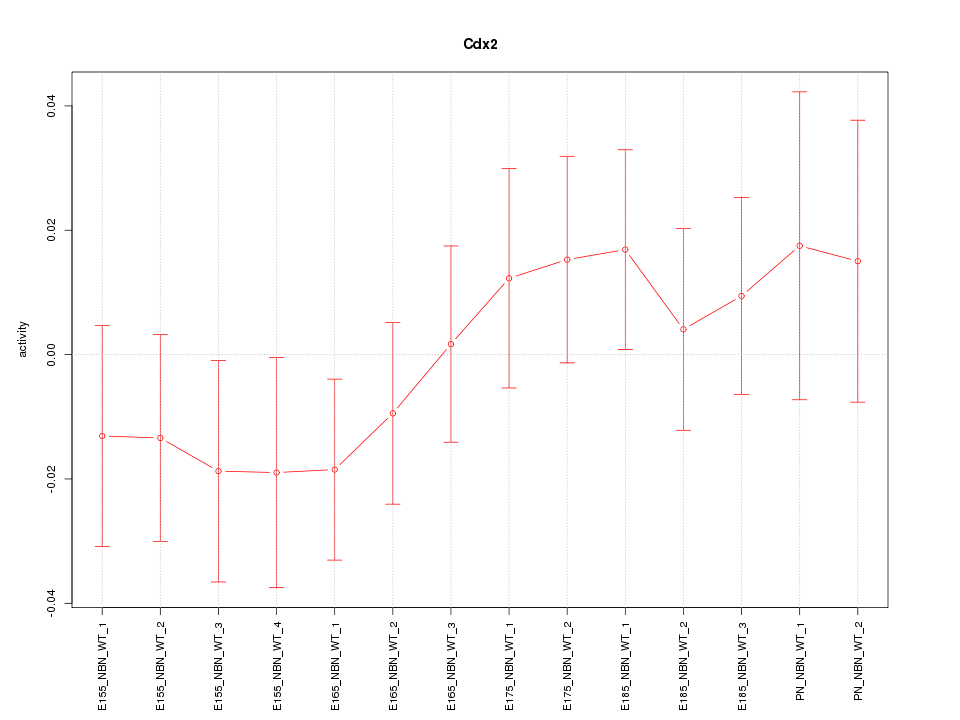
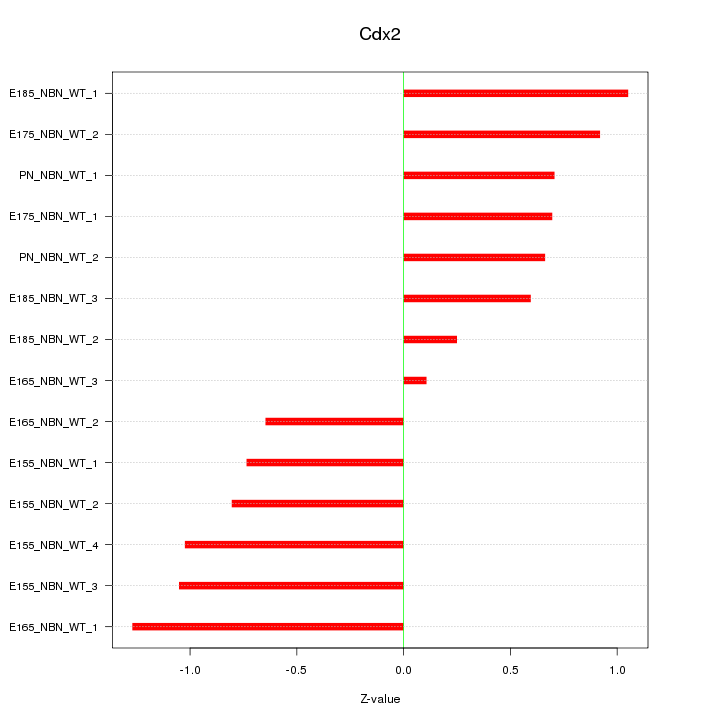
| 0.8 |
2.5 |
GO:0048818 |
positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.7 |
2.9 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.6 |
1.8 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.4 |
3.0 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.2 |
1.1 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.2 |
1.9 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 |
1.3 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.2 |
2.2 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.2 |
0.9 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 |
2.6 |
GO:0035994 |
response to muscle stretch(GO:0035994) |
| 0.1 |
0.5 |
GO:0021856 |
cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 |
0.5 |
GO:0021555 |
midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 |
0.5 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.2 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.0 |
1.0 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.7 |
GO:0032291 |
central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 |
0.2 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.0 |
0.5 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 |
0.7 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.2 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.1 |
GO:0071285 |
cellular response to lithium ion(GO:0071285) |
| 0.0 |
0.2 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.6 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.6 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.0 |
0.2 |
GO:0019369 |
drug metabolic process(GO:0017144) arachidonic acid metabolic process(GO:0019369) |
| 0.0 |
0.1 |
GO:0048664 |
neuron fate determination(GO:0048664) |