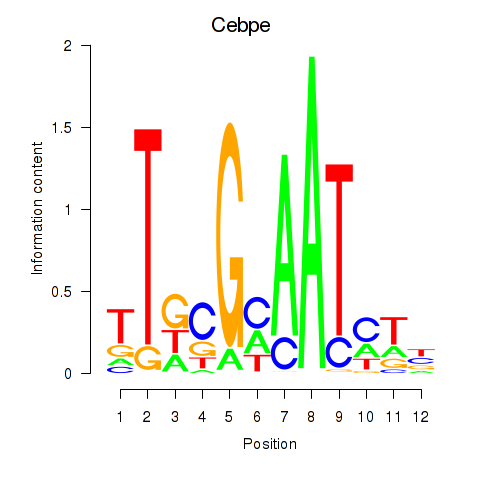
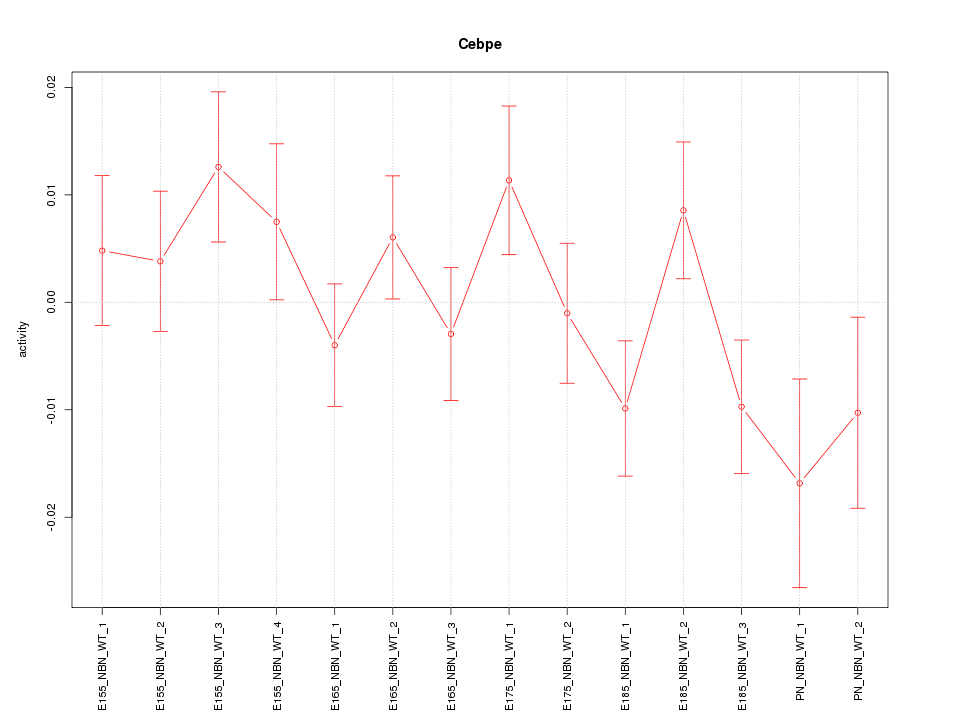
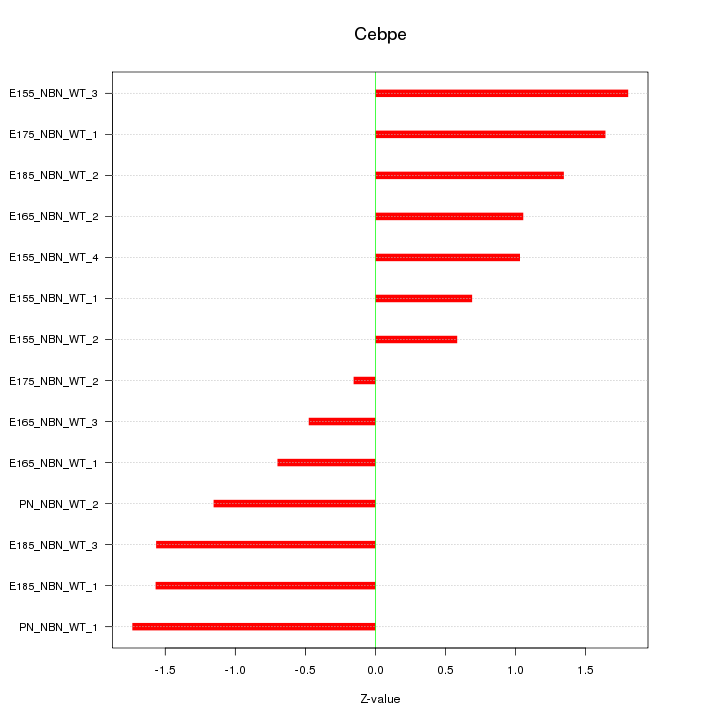
| 0.6 |
3.0 |
GO:0002121 |
inter-male aggressive behavior(GO:0002121) |
| 0.4 |
1.2 |
GO:1905065 |
hematopoietic stem cell migration(GO:0035701) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.4 |
1.6 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.4 |
1.5 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.4 |
0.8 |
GO:2000338 |
chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.3 |
2.4 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.3 |
1.0 |
GO:0045410 |
positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.3 |
1.3 |
GO:0032382 |
positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.3 |
1.5 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.3 |
0.8 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.3 |
2.1 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 |
0.9 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 0.2 |
1.4 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.2 |
3.6 |
GO:1901841 |
regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.2 |
0.9 |
GO:0061526 |
acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.2 |
1.1 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.2 |
0.7 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 |
0.5 |
GO:0030862 |
neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.2 |
1.6 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.2 |
0.6 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.2 |
0.8 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 |
0.5 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 |
0.6 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 |
0.4 |
GO:0010751 |
regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 |
0.4 |
GO:0046168 |
glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 |
0.5 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.1 |
0.5 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.1 |
1.1 |
GO:1902669 |
positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.1 |
2.3 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
0.7 |
GO:0001927 |
exocyst assembly(GO:0001927) |
| 0.1 |
0.7 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 |
0.6 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 |
0.4 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 |
0.5 |
GO:0051643 |
endoplasmic reticulum localization(GO:0051643) |
| 0.1 |
1.8 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.1 |
0.3 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 |
0.9 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 |
0.8 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.1 |
0.3 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.1 |
0.4 |
GO:1901491 |
negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 |
0.3 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.1 |
0.3 |
GO:1900369 |
negative regulation of RNA interference(GO:1900369) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 |
0.5 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 |
0.4 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.1 |
0.6 |
GO:0051790 |
short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 |
0.2 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.1 |
0.3 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.1 |
0.5 |
GO:0003105 |
negative regulation of glomerular filtration(GO:0003105) |
| 0.1 |
0.5 |
GO:0060467 |
negative regulation of fertilization(GO:0060467) |
| 0.1 |
1.6 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.1 |
0.3 |
GO:0048818 |
embryonic nail plate morphogenesis(GO:0035880) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.1 |
0.3 |
GO:0002925 |
positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 |
0.6 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 |
0.3 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 |
0.3 |
GO:1903722 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) regulation of centriole elongation(GO:1903722) |
| 0.1 |
0.9 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 |
3.4 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 |
0.3 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.1 |
0.3 |
GO:0034239 |
regulation of macrophage fusion(GO:0034239) |
| 0.1 |
0.6 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.1 |
0.7 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 |
0.4 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.1 |
0.6 |
GO:0034433 |
steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 |
0.2 |
GO:0048352 |
neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.1 |
0.3 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.1 |
0.2 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 |
0.4 |
GO:0016078 |
tRNA catabolic process(GO:0016078) |
| 0.1 |
0.4 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 |
0.2 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 |
0.4 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 |
0.4 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 |
0.6 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.3 |
GO:1901740 |
negative regulation of myoblast fusion(GO:1901740) |
| 0.1 |
0.2 |
GO:0046381 |
CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.1 |
0.7 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.3 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.1 |
0.3 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 |
0.1 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 |
0.3 |
GO:1903898 |
positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 |
1.3 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.1 |
0.3 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 |
0.7 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.2 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 |
0.3 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 |
0.8 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.1 |
0.2 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 |
0.8 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.1 |
0.6 |
GO:0035562 |
negative regulation of chromatin binding(GO:0035562) |
| 0.1 |
0.2 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.1 |
0.7 |
GO:0002097 |
tRNA wobble base modification(GO:0002097) |
| 0.1 |
0.3 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.1 |
0.2 |
GO:0097309 |
cap1 mRNA methylation(GO:0097309) |
| 0.1 |
0.4 |
GO:1903441 |
protein localization to ciliary membrane(GO:1903441) |
| 0.1 |
0.4 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.1 |
0.2 |
GO:0018158 |
protein oxidation(GO:0018158) |
| 0.1 |
0.6 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
1.4 |
GO:0000460 |
maturation of 5.8S rRNA(GO:0000460) |
| 0.0 |
0.1 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.0 |
0.4 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.0 |
0.1 |
GO:0019255 |
glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 |
0.2 |
GO:0048539 |
mature ribosome assembly(GO:0042256) bone marrow development(GO:0048539) |
| 0.0 |
0.0 |
GO:0006701 |
progesterone biosynthetic process(GO:0006701) |
| 0.0 |
0.6 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.0 |
0.8 |
GO:0052646 |
alditol phosphate metabolic process(GO:0052646) |
| 0.0 |
0.2 |
GO:0022410 |
circadian sleep/wake cycle process(GO:0022410) regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) circadian sleep/wake cycle, sleep(GO:0050802) |
| 0.0 |
0.2 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.0 |
0.1 |
GO:0060282 |
positive regulation of oocyte development(GO:0060282) |
| 0.0 |
0.5 |
GO:0070498 |
interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 |
0.1 |
GO:0006990 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 |
1.7 |
GO:0048821 |
erythrocyte development(GO:0048821) |
| 0.0 |
0.1 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.0 |
0.7 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
0.2 |
GO:0018364 |
peptidyl-glutamine methylation(GO:0018364) snoRNA 3'-end processing(GO:0031126) |
| 0.0 |
0.1 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 |
1.2 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 |
0.1 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.0 |
0.2 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 |
0.3 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.0 |
0.1 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.0 |
0.2 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 |
0.1 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.0 |
0.1 |
GO:0046294 |
formaldehyde catabolic process(GO:0046294) |
| 0.0 |
1.7 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
0.2 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 |
0.2 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.4 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.0 |
0.3 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 |
0.4 |
GO:0097066 |
response to thyroid hormone(GO:0097066) |
| 0.0 |
0.1 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.1 |
GO:0033860 |
regulation of NAD(P)H oxidase activity(GO:0033860) flavin-containing compound metabolic process(GO:0042726) |
| 0.0 |
0.4 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
1.3 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.0 |
0.2 |
GO:0060576 |
intestinal epithelial cell development(GO:0060576) |
| 0.0 |
0.0 |
GO:0070782 |
phospholipid scrambling(GO:0017121) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) regulation of membrane lipid distribution(GO:0097035) |
| 0.0 |
0.6 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 |
0.3 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 |
0.1 |
GO:0014835 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 |
0.2 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.2 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.0 |
0.1 |
GO:0030647 |
polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 |
0.1 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.0 |
0.2 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
0.3 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 |
0.2 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.1 |
GO:0046032 |
ADP catabolic process(GO:0046032) |
| 0.0 |
0.2 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 |
1.1 |
GO:0001947 |
heart looping(GO:0001947) |
| 0.0 |
0.2 |
GO:0044791 |
modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 |
0.7 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.1 |
GO:2000767 |
positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 |
0.3 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.5 |
GO:0006110 |
regulation of glycolytic process(GO:0006110) |
| 0.0 |
0.2 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.2 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) |
| 0.0 |
0.2 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.1 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.0 |
0.4 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 |
0.2 |
GO:0001956 |
positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.0 |
0.5 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.5 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.0 |
0.2 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.2 |
GO:0030432 |
peristalsis(GO:0030432) |
| 0.0 |
0.1 |
GO:0070301 |
cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 |
0.7 |
GO:0022401 |
desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.0 |
0.3 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.2 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.0 |
0.1 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 |
0.3 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.2 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.4 |
GO:2000369 |
regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 |
0.5 |
GO:0009303 |
rRNA transcription(GO:0009303) |
| 0.0 |
0.2 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.2 |
GO:0019348 |
polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 |
0.1 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.0 |
0.1 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.0 |
0.2 |
GO:1903624 |
regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.0 |
0.8 |
GO:0050982 |
detection of mechanical stimulus(GO:0050982) |
| 0.0 |
0.5 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.5 |
GO:0045760 |
positive regulation of action potential(GO:0045760) |
| 0.0 |
0.2 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 |
0.1 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 |
0.6 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.2 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 |
0.1 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 |
0.2 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.0 |
0.1 |
GO:0051490 |
negative regulation of filopodium assembly(GO:0051490) |
| 0.0 |
0.2 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.5 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
0.8 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.0 |
0.3 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.1 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.0 |
1.4 |
GO:0060968 |
regulation of gene silencing(GO:0060968) |
| 0.0 |
0.1 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.0 |
0.3 |
GO:0044872 |
lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 |
0.1 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.0 |
0.3 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
4.3 |
GO:0006869 |
lipid transport(GO:0006869) |
| 0.0 |
0.1 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.0 |
0.3 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 |
0.1 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.4 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.2 |
GO:0009186 |
deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 |
0.3 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.2 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.6 |
GO:0045761 |
regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 |
0.4 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 |
0.3 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.1 |
GO:1902224 |
cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.0 |
0.2 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.2 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.4 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
0.2 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.2 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.1 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.0 |
0.2 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 |
0.2 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.1 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.0 |
0.9 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.8 |
GO:0060395 |
SMAD protein signal transduction(GO:0060395) |
| 0.0 |
0.1 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.1 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.3 |
GO:0090280 |
positive regulation of calcium ion import(GO:0090280) |
| 0.0 |
0.0 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 |
0.1 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 |
0.1 |
GO:0015669 |
gas transport(GO:0015669) |
| 0.0 |
0.2 |
GO:0010667 |
negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 |
0.2 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.2 |
GO:0007289 |
spermatid nucleus differentiation(GO:0007289) |
| 0.0 |
0.1 |
GO:0070126 |
mitochondrial translational termination(GO:0070126) |
| 0.0 |
0.3 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 |
0.1 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.2 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.3 |
GO:0048745 |
smooth muscle tissue development(GO:0048745) |
| 0.0 |
0.1 |
GO:0010666 |
positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 |
0.1 |
GO:0033152 |
immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 |
0.1 |
GO:0010992 |
ubiquitin homeostasis(GO:0010992) |