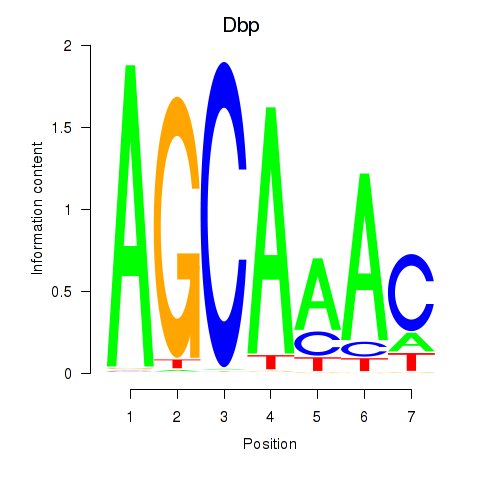
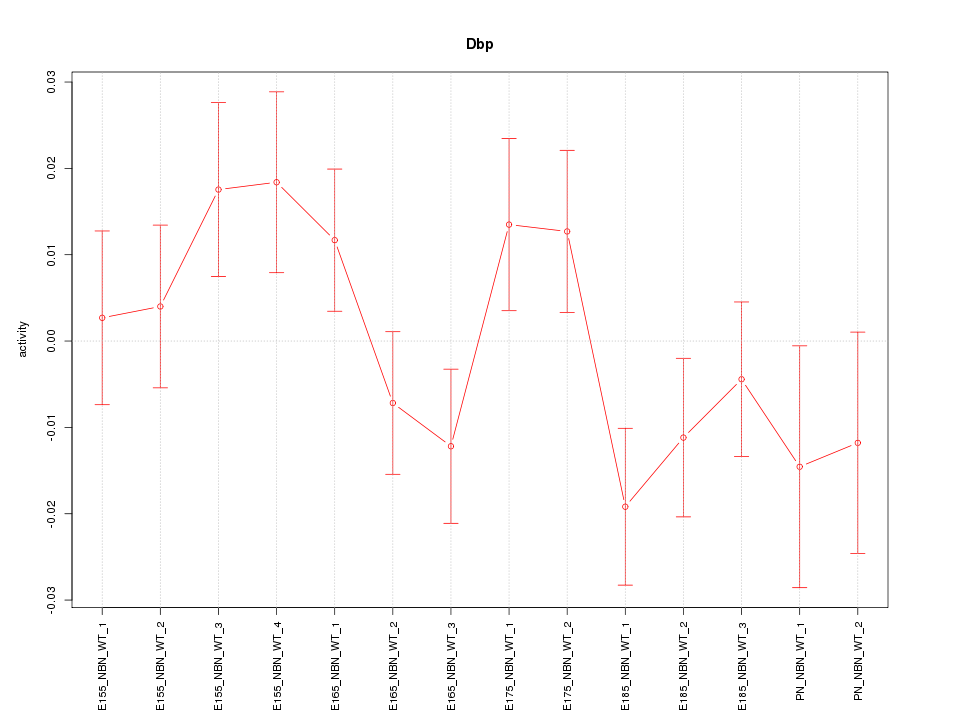
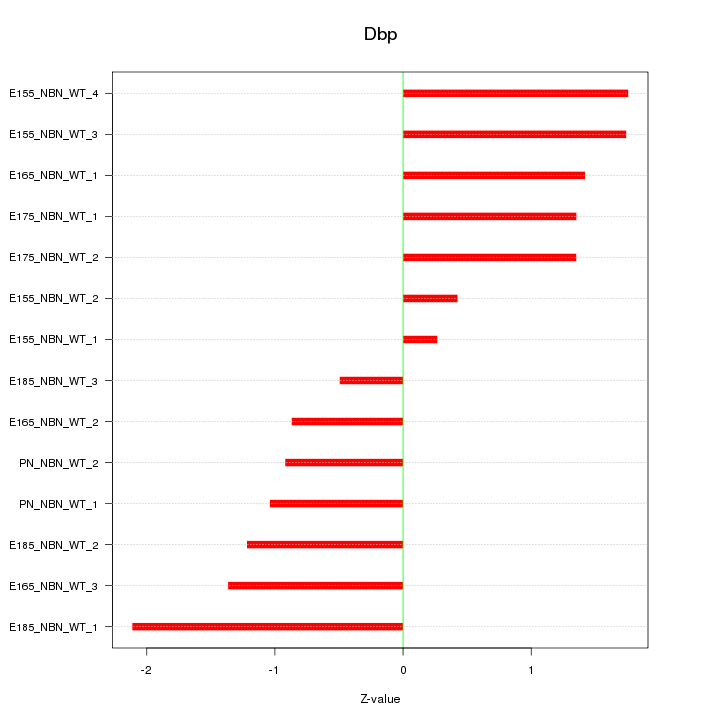
| 1.5 |
4.5 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.6 |
3.7 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.6 |
1.7 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.5 |
2.5 |
GO:2000302 |
regulation of synaptic vesicle endocytosis(GO:1900242) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.5 |
1.9 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.5 |
3.7 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.4 |
6.7 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.4 |
1.3 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.4 |
3.4 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.4 |
1.2 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.4 |
1.5 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.3 |
1.0 |
GO:0003219 |
cardiac right ventricle formation(GO:0003219) |
| 0.3 |
3.6 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.3 |
0.9 |
GO:0021852 |
pyramidal neuron migration(GO:0021852) |
| 0.3 |
1.7 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.3 |
0.8 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 |
0.7 |
GO:0060084 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 0.2 |
2.1 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.2 |
2.3 |
GO:0014824 |
artery smooth muscle contraction(GO:0014824) |
| 0.2 |
0.7 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.2 |
1.3 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 |
0.9 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.2 |
1.2 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 |
2.4 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 |
0.7 |
GO:0002072 |
optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.2 |
1.4 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.2 |
0.8 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.2 |
0.8 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.2 |
3.3 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.2 |
0.5 |
GO:0003289 |
atrial septum primum morphogenesis(GO:0003289) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 |
0.4 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 |
0.6 |
GO:2001045 |
closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 |
0.6 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 |
0.5 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 0.1 |
0.4 |
GO:2000224 |
testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 |
0.9 |
GO:0071361 |
cellular response to zinc ion(GO:0071294) cellular response to ethanol(GO:0071361) |
| 0.1 |
0.4 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) cerebellum vasculature development(GO:0061300) |
| 0.1 |
0.3 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.1 |
0.2 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.8 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 |
1.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
0.8 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 |
2.3 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 |
0.9 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
0.4 |
GO:0010993 |
regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 |
1.0 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.4 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 |
0.3 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
2.2 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 |
0.2 |
GO:2000170 |
positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.1 |
0.9 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 |
1.4 |
GO:0021702 |
cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 |
1.3 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
0.6 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
4.9 |
GO:0060395 |
SMAD protein signal transduction(GO:0060395) |
| 0.1 |
0.4 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
1.4 |
GO:0010669 |
epithelial structure maintenance(GO:0010669) |
| 0.1 |
0.1 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 |
1.9 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
0.8 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
1.4 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.1 |
0.2 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.1 |
0.3 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.5 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 |
0.2 |
GO:0097105 |
presynaptic membrane assembly(GO:0097105) |
| 0.0 |
0.5 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
3.4 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.5 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 |
3.3 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.5 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.2 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.6 |
GO:2000052 |
positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 |
0.2 |
GO:1902965 |
regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 |
0.1 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.0 |
0.7 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.7 |
GO:0071425 |
hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 |
0.5 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.4 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.0 |
0.5 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.1 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.0 |
0.1 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.0 |
0.2 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.4 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.1 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 |
1.7 |
GO:0006487 |
protein N-linked glycosylation(GO:0006487) |
| 0.0 |
0.5 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.9 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
1.0 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 |
0.3 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.2 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.5 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.2 |
GO:0021540 |
corpus callosum morphogenesis(GO:0021540) |
| 0.0 |
0.1 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.3 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.2 |
GO:0030007 |
cellular potassium ion homeostasis(GO:0030007) |
| 0.0 |
0.1 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.0 |
1.1 |
GO:0003073 |
regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 |
0.2 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
0.3 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
0.3 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.5 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.1 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 |
0.1 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 |
0.2 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.7 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.3 |
GO:0007131 |
reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |