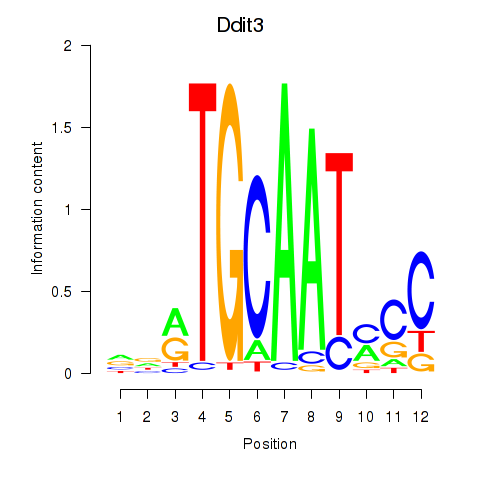
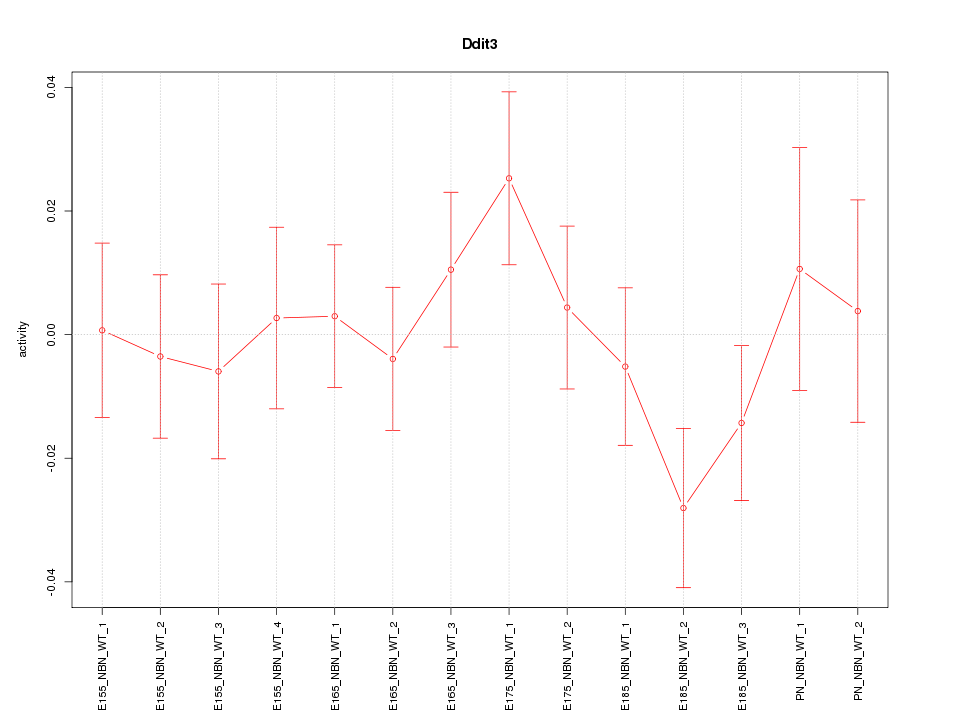
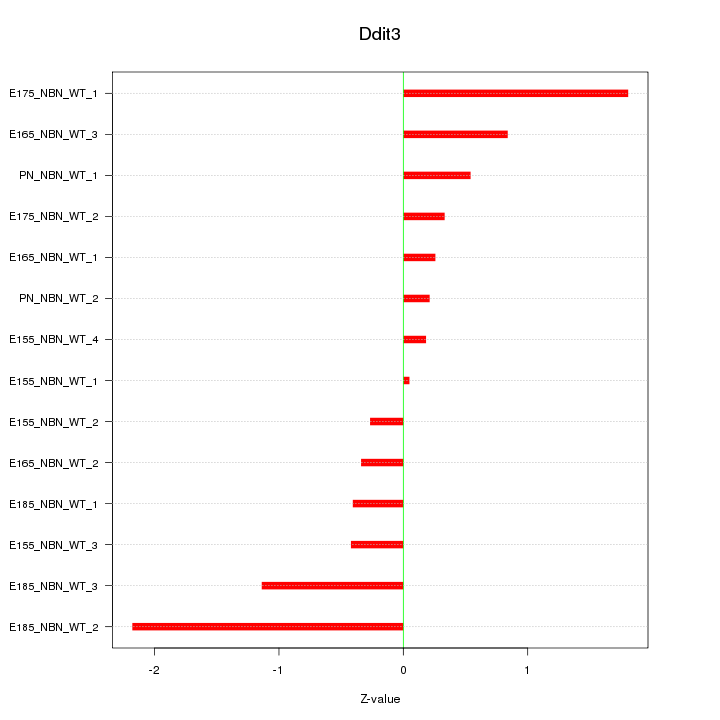
| 0.3 |
3.0 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 |
0.6 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 |
0.6 |
GO:0070649 |
meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 |
0.5 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 |
0.6 |
GO:0050748 |
negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.1 |
0.4 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 |
0.7 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 |
0.3 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 |
0.3 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 |
0.3 |
GO:0098904 |
regulation of AV node cell action potential(GO:0098904) |
| 0.1 |
0.6 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.1 |
0.2 |
GO:0034334 |
adherens junction maintenance(GO:0034334) |
| 0.1 |
0.5 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 |
0.3 |
GO:0071205 |
clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 |
0.4 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.1 |
0.3 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.1 |
0.2 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 0.1 |
0.4 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.2 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.1 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.0 |
0.2 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.5 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 |
0.2 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 |
0.3 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.1 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 |
0.2 |
GO:0010747 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 |
0.3 |
GO:0016139 |
glycoside catabolic process(GO:0016139) |
| 0.0 |
0.2 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.0 |
0.2 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 |
0.1 |
GO:0040038 |
meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 |
0.1 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 0.0 |
0.2 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.1 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 |
0.3 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.0 |
0.4 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.6 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.0 |
0.8 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.5 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.0 |
0.3 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.1 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 |
0.4 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.2 |
GO:0006273 |
lagging strand elongation(GO:0006273) |
| 0.0 |
0.1 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 |
0.1 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.0 |
0.5 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.5 |
GO:0031571 |
mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 |
0.1 |
GO:2000973 |
regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 |
0.3 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.3 |
GO:0010960 |
magnesium ion homeostasis(GO:0010960) |
| 0.0 |
0.0 |
GO:0038027 |
apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.0 |
0.1 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.1 |
GO:0036089 |
cleavage furrow formation(GO:0036089) |
| 0.0 |
0.0 |
GO:0042275 |
error-free postreplication DNA repair(GO:0042275) |
| 0.0 |
0.2 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |