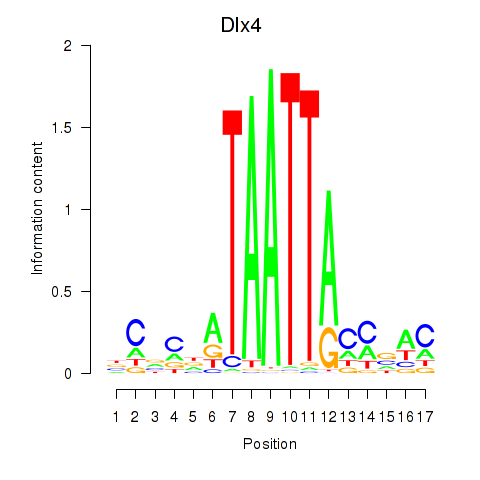
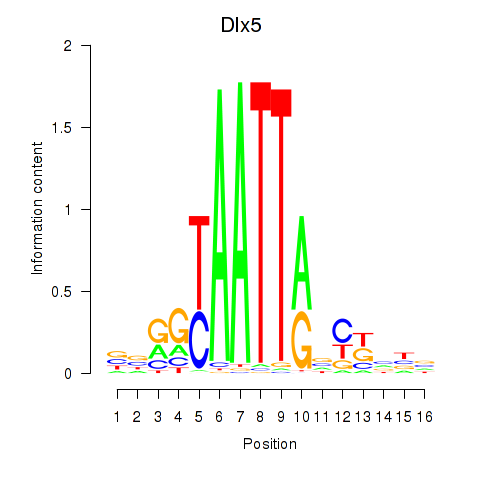
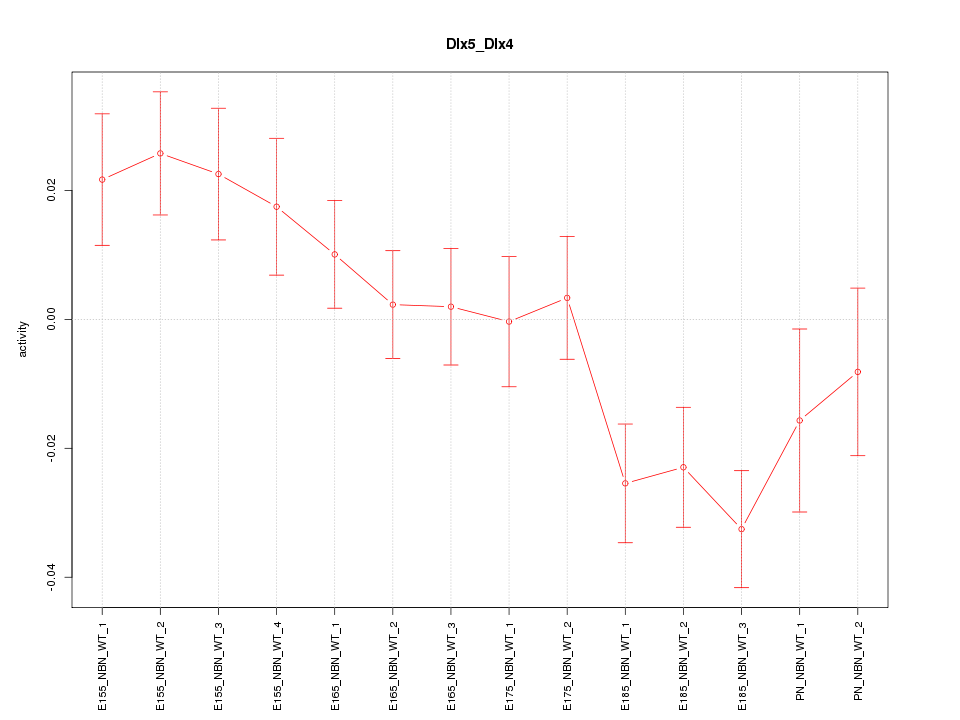
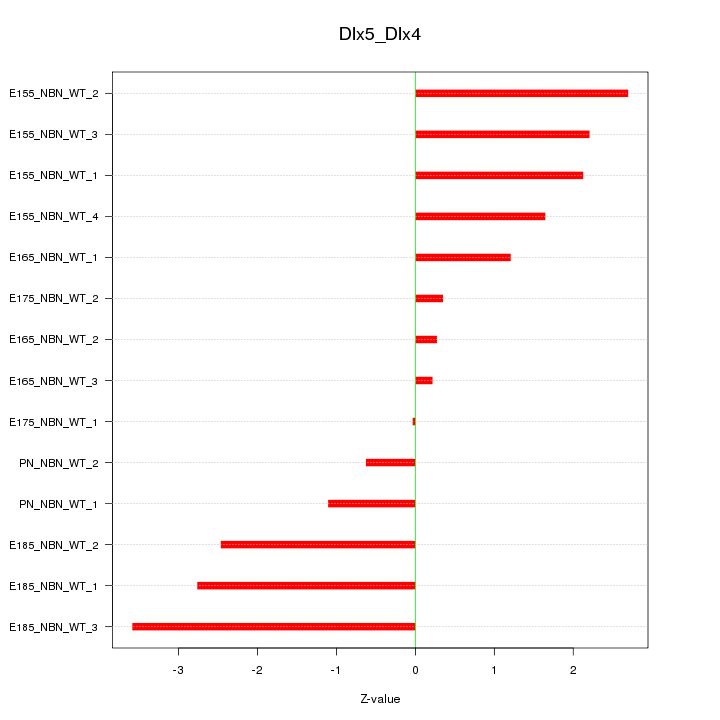
| 2.1 |
6.2 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 2.0 |
9.9 |
GO:0002121 |
inter-male aggressive behavior(GO:0002121) |
| 1.8 |
5.4 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 1.7 |
10.3 |
GO:0046549 |
retinal cone cell development(GO:0046549) |
| 1.1 |
3.4 |
GO:0038165 |
oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.9 |
2.7 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.8 |
3.0 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.6 |
1.7 |
GO:1904884 |
telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.5 |
2.1 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.5 |
1.6 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.5 |
1.5 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.5 |
2.9 |
GO:0051790 |
short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.4 |
1.3 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.4 |
1.7 |
GO:0070885 |
negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) cellular response to copper ion(GO:0071280) |
| 0.4 |
4.1 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.4 |
1.6 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.4 |
2.0 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.4 |
1.1 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.3 |
1.4 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.3 |
1.0 |
GO:0070649 |
meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.3 |
1.0 |
GO:0070212 |
protein poly-ADP-ribosylation(GO:0070212) |
| 0.3 |
1.0 |
GO:0061300 |
progesterone secretion(GO:0042701) cerebellum vasculature development(GO:0061300) |
| 0.3 |
2.3 |
GO:0097151 |
positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.3 |
2.4 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.3 |
8.2 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.3 |
0.8 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.3 |
0.8 |
GO:2000481 |
positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.3 |
1.0 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.2 |
1.2 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.2 |
1.9 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 |
1.4 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 |
3.1 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 |
1.1 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.2 |
0.4 |
GO:1904339 |
negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.2 |
1.2 |
GO:0002326 |
B cell lineage commitment(GO:0002326) ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 |
0.6 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.2 |
1.8 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.2 |
0.6 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.2 |
0.6 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 |
1.3 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.2 |
0.2 |
GO:0097694 |
establishment of RNA localization to telomere(GO:0097694) |
| 0.2 |
1.1 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 |
1.4 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 |
1.0 |
GO:1903056 |
melanocyte migration(GO:0097324) regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 |
0.7 |
GO:0014829 |
vascular smooth muscle contraction(GO:0014829) |
| 0.2 |
1.0 |
GO:0048840 |
otolith development(GO:0048840) |
| 0.2 |
0.5 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 |
0.5 |
GO:0006404 |
RNA import into nucleus(GO:0006404) snRNA transport(GO:0051030) |
| 0.2 |
0.8 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.2 |
2.9 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.2 |
0.5 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.2 |
1.1 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.2 |
0.9 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.1 |
0.6 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.1 |
0.9 |
GO:0060527 |
prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.1 |
2.4 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.1 |
0.4 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 |
0.7 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 |
2.1 |
GO:0048875 |
chemical homeostasis within a tissue(GO:0048875) |
| 0.1 |
1.5 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 |
0.4 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 |
0.7 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.1 |
0.7 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.1 |
0.4 |
GO:0098501 |
polynucleotide dephosphorylation(GO:0098501) |
| 0.1 |
0.4 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 |
0.4 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 |
1.3 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.1 |
5.9 |
GO:0008347 |
glial cell migration(GO:0008347) |
| 0.1 |
0.4 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 |
0.9 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
1.0 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.1 |
0.2 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 |
1.5 |
GO:0034390 |
smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 |
3.0 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
0.6 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
2.8 |
GO:0006099 |
tricarboxylic acid cycle(GO:0006099) |
| 0.1 |
0.4 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 |
0.9 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
1.9 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.1 |
0.3 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.1 |
0.7 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.1 |
0.8 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 |
0.5 |
GO:0045583 |
regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 |
0.3 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.1 |
0.3 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 |
0.7 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
0.6 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 |
0.2 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 |
0.3 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 |
0.2 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) |
| 0.1 |
0.5 |
GO:2001012 |
mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 |
0.2 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.1 |
0.4 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.1 |
0.4 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 |
1.6 |
GO:0003009 |
skeletal muscle contraction(GO:0003009) |
| 0.1 |
0.3 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.1 |
0.3 |
GO:0030647 |
polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.1 |
0.2 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 |
0.2 |
GO:1901856 |
negative regulation of cellular respiration(GO:1901856) |
| 0.1 |
0.4 |
GO:0060768 |
regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 |
0.3 |
GO:1903546 |
protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 |
0.2 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 |
0.6 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.1 |
0.1 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 |
0.2 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.1 |
0.3 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.1 |
0.2 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 |
0.7 |
GO:0032570 |
response to progesterone(GO:0032570) |
| 0.1 |
0.6 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 |
0.2 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 |
1.4 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 |
1.8 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.1 |
0.2 |
GO:0018158 |
protein oxidation(GO:0018158) |
| 0.1 |
0.3 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.1 |
0.9 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.1 |
0.2 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.1 |
0.4 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.1 |
0.2 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.1 |
1.1 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.1 |
1.3 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 |
0.5 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 |
0.1 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 0.1 |
1.4 |
GO:0072337 |
modified amino acid transport(GO:0072337) |
| 0.1 |
0.6 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 |
0.8 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.0 |
0.2 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 |
0.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.3 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.0 |
0.2 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.0 |
0.3 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.0 |
1.1 |
GO:0003016 |
respiratory system process(GO:0003016) |
| 0.0 |
0.8 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.6 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.5 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.2 |
GO:0060398 |
regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 |
0.1 |
GO:0032490 |
detection of molecule of bacterial origin(GO:0032490) |
| 0.0 |
0.1 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.0 |
0.5 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
3.1 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.2 |
GO:0061025 |
membrane fusion(GO:0061025) |
| 0.0 |
0.1 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.0 |
0.7 |
GO:0003417 |
growth plate cartilage development(GO:0003417) |
| 0.0 |
0.1 |
GO:0000965 |
mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 |
0.2 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 |
0.4 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.2 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.3 |
GO:0000467 |
exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 |
0.5 |
GO:0071624 |
positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 |
0.9 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.0 |
0.3 |
GO:0043206 |
extracellular fibril organization(GO:0043206) |
| 0.0 |
0.5 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 |
0.3 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
0.2 |
GO:0051497 |
negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 |
0.4 |
GO:0033005 |
positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
1.5 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
0.8 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
1.2 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.4 |
GO:0035094 |
response to nicotine(GO:0035094) |
| 0.0 |
0.3 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
1.2 |
GO:0050982 |
detection of mechanical stimulus(GO:0050982) |
| 0.0 |
0.5 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.2 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 |
0.2 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.0 |
0.1 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 |
0.4 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.1 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 |
1.4 |
GO:0002224 |
toll-like receptor signaling pathway(GO:0002224) |
| 0.0 |
0.1 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.0 |
0.9 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.0 |
1.7 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.2 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 |
3.4 |
GO:0006261 |
DNA-dependent DNA replication(GO:0006261) |
| 0.0 |
0.3 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.3 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 |
0.1 |
GO:0060065 |
uterus development(GO:0060065) |
| 0.0 |
0.1 |
GO:0035457 |
cellular response to interferon-alpha(GO:0035457) |
| 0.0 |
1.9 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.3 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.0 |
1.0 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 |
0.2 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.0 |
0.4 |
GO:0040019 |
positive regulation of embryonic development(GO:0040019) |
| 0.0 |
0.3 |
GO:0019730 |
antimicrobial humoral response(GO:0019730) |
| 0.0 |
0.1 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 |
0.8 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 |
0.1 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.9 |
GO:0051438 |
regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 |
0.4 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.0 |
0.2 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.0 |
0.5 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
0.2 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.0 |
0.2 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.3 |
GO:1903846 |
positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 |
0.6 |
GO:0051647 |
nucleus localization(GO:0051647) |
| 0.0 |
0.1 |
GO:1900039 |
positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 |
2.8 |
GO:0043062 |
extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 |
0.2 |
GO:0060746 |
parental behavior(GO:0060746) |
| 0.0 |
0.1 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 |
0.0 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 |
2.5 |
GO:0007266 |
Rho protein signal transduction(GO:0007266) |
| 0.0 |
0.1 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.4 |
GO:0045727 |
positive regulation of cellular amide metabolic process(GO:0034250) positive regulation of translation(GO:0045727) |
| 0.0 |
0.2 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.3 |
GO:0000578 |
embryonic axis specification(GO:0000578) |
| 0.0 |
0.0 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.0 |
0.4 |
GO:0001947 |
heart looping(GO:0001947) |
| 0.0 |
0.4 |
GO:0001523 |
retinoid metabolic process(GO:0001523) |
| 0.0 |
0.3 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.5 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.0 |
0.3 |
GO:0035315 |
hair cell differentiation(GO:0035315) auditory receptor cell differentiation(GO:0042491) |
| 0.0 |
0.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.5 |
GO:0003073 |
regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 |
0.1 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.3 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
1.3 |
GO:0000077 |
DNA damage checkpoint(GO:0000077) DNA integrity checkpoint(GO:0031570) |
| 0.0 |
0.1 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.3 |
GO:0032781 |
positive regulation of ATPase activity(GO:0032781) |
| 0.0 |
0.4 |
GO:0001541 |
ovarian follicle development(GO:0001541) |
| 0.0 |
0.4 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.4 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.5 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.5 |
GO:0043407 |
negative regulation of MAP kinase activity(GO:0043407) |