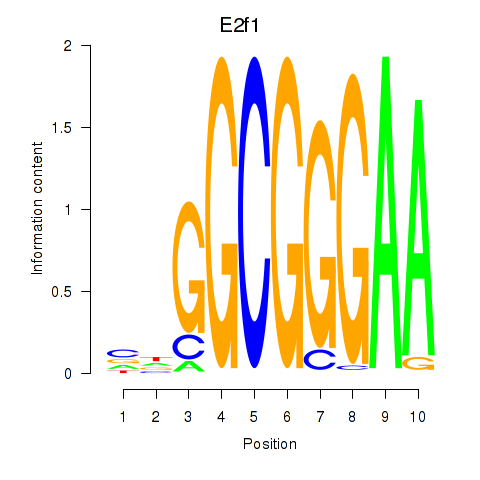
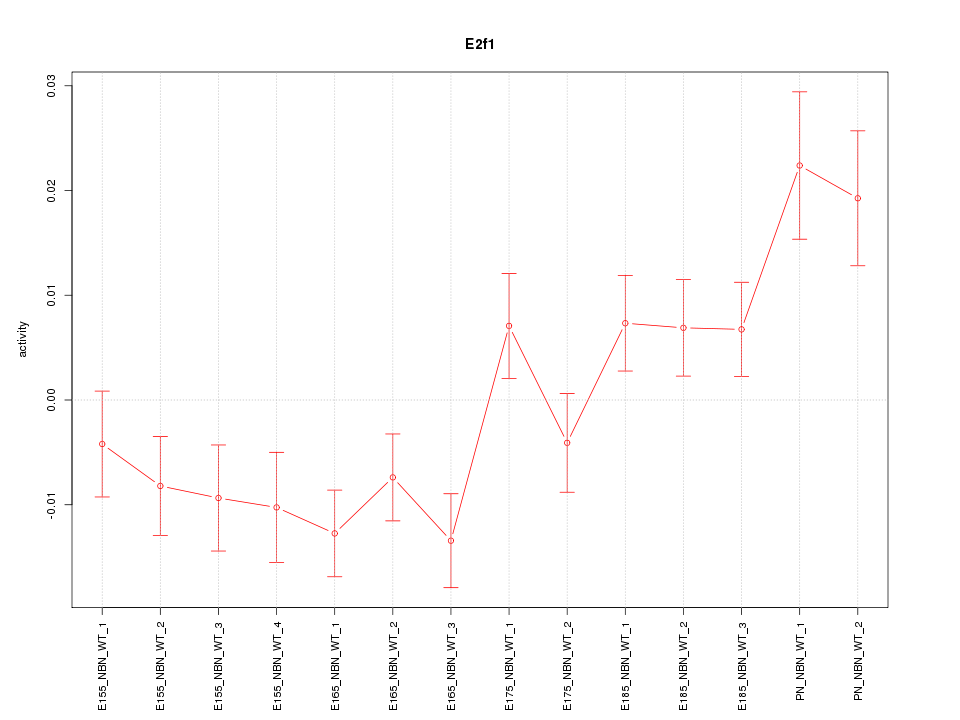
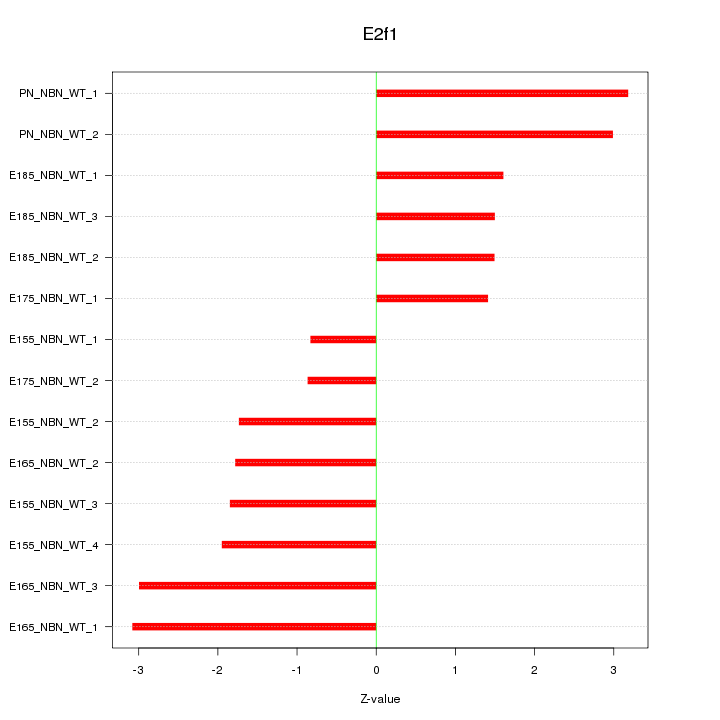
| 1.9 |
5.7 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.7 |
7.0 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 1.6 |
1.6 |
GO:0002432 |
granuloma formation(GO:0002432) |
| 1.5 |
5.8 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.2 |
3.7 |
GO:1902630 |
regulation of membrane hyperpolarization(GO:1902630) |
| 1.2 |
3.6 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 1.2 |
3.5 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 1.0 |
3.0 |
GO:0021837 |
motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.9 |
2.7 |
GO:0072180 |
mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) |
| 0.9 |
0.9 |
GO:0007418 |
ventral midline development(GO:0007418) |
| 0.9 |
2.6 |
GO:1901254 |
positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.9 |
3.5 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.9 |
3.4 |
GO:0048143 |
astrocyte activation(GO:0048143) |
| 0.8 |
3.3 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.8 |
5.6 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.8 |
3.1 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.7 |
5.6 |
GO:0002347 |
response to tumor cell(GO:0002347) |
| 0.7 |
4.2 |
GO:0035989 |
tendon development(GO:0035989) |
| 0.7 |
2.0 |
GO:0033860 |
regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.7 |
2.0 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) pancreas morphogenesis(GO:0061113) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.6 |
1.9 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.6 |
1.9 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.6 |
1.2 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.6 |
1.8 |
GO:0072070 |
loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.6 |
1.8 |
GO:0051026 |
chiasma assembly(GO:0051026) |
| 0.6 |
2.4 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.6 |
2.9 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.6 |
2.9 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.6 |
1.7 |
GO:0043465 |
regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.6 |
1.7 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.6 |
2.3 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.5 |
2.7 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.5 |
2.7 |
GO:0046601 |
positive regulation of centriole replication(GO:0046601) |
| 0.5 |
1.5 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.5 |
2.9 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.5 |
4.7 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.5 |
7.0 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.5 |
0.5 |
GO:0002034 |
angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.4 |
1.7 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.4 |
0.9 |
GO:0045799 |
positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.4 |
0.8 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.4 |
1.2 |
GO:0051464 |
positive regulation of cortisol secretion(GO:0051464) |
| 0.4 |
1.2 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 0.4 |
1.2 |
GO:0010957 |
negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.4 |
1.1 |
GO:1900158 |
regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 |
2.9 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.4 |
1.5 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) |
| 0.3 |
1.7 |
GO:0042891 |
antibiotic transport(GO:0042891) |
| 0.3 |
1.0 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.3 |
1.4 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.3 |
1.7 |
GO:0021905 |
pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) |
| 0.3 |
1.4 |
GO:0060266 |
positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.3 |
0.3 |
GO:0060423 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.3 |
1.3 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.3 |
1.0 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 |
3.5 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.3 |
0.9 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.3 |
1.9 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.3 |
0.9 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.3 |
1.2 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.3 |
0.9 |
GO:2000620 |
programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 |
1.5 |
GO:0042997 |
negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.3 |
2.3 |
GO:0033504 |
floor plate development(GO:0033504) |
| 0.3 |
0.9 |
GO:0034184 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.3 |
1.4 |
GO:0032298 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.3 |
1.1 |
GO:0032417 |
positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.3 |
0.8 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 0.3 |
1.4 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) cellular response to potassium ion starvation(GO:0051365) |
| 0.3 |
1.4 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.3 |
0.8 |
GO:1902498 |
regulation of protein autoubiquitination(GO:1902498) |
| 0.3 |
3.5 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.3 |
1.3 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.3 |
1.6 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.3 |
1.6 |
GO:0098789 |
pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 |
1.6 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.3 |
0.8 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.3 |
3.5 |
GO:0044854 |
plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) |
| 0.3 |
1.1 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.3 |
0.8 |
GO:0097402 |
neuroblast migration(GO:0097402) |
| 0.2 |
1.2 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 |
1.0 |
GO:1903519 |
apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.2 |
4.4 |
GO:0021516 |
dorsal spinal cord development(GO:0021516) |
| 0.2 |
3.9 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.2 |
1.4 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 |
0.5 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.2 |
1.6 |
GO:0010273 |
detoxification of copper ion(GO:0010273) detoxification of inorganic compound(GO:0061687) cellular response to zinc ion(GO:0071294) stress response to copper ion(GO:1990169) |
| 0.2 |
1.2 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 |
1.2 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 |
0.5 |
GO:2000670 |
positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.2 |
1.1 |
GO:0072383 |
plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.2 |
0.7 |
GO:0019043 |
establishment of viral latency(GO:0019043) |
| 0.2 |
0.9 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 |
14.5 |
GO:0035019 |
somatic stem cell population maintenance(GO:0035019) |
| 0.2 |
1.5 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.2 |
1.5 |
GO:1901162 |
serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.2 |
1.1 |
GO:0051315 |
attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 |
1.5 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.2 |
1.7 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.2 |
0.4 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.2 |
3.9 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.2 |
1.7 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.2 |
0.6 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.2 |
1.6 |
GO:2000653 |
regulation of genetic imprinting(GO:2000653) |
| 0.2 |
0.6 |
GO:0046655 |
folic acid metabolic process(GO:0046655) |
| 0.2 |
5.1 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.2 |
1.0 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 |
0.2 |
GO:1903525 |
regulation of membrane tubulation(GO:1903525) |
| 0.2 |
0.6 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.2 |
0.6 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.2 |
2.6 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.2 |
0.4 |
GO:2000418 |
positive regulation of eosinophil migration(GO:2000418) |
| 0.2 |
0.8 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.2 |
1.1 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 |
0.5 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.2 |
0.5 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.2 |
3.1 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.2 |
0.9 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.2 |
1.0 |
GO:0032815 |
negative regulation of natural killer cell activation(GO:0032815) |
| 0.2 |
0.5 |
GO:0046078 |
deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) dUMP metabolic process(GO:0046078) |
| 0.2 |
0.5 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 |
1.8 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 |
0.8 |
GO:0060754 |
positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 |
0.3 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 |
0.5 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.2 |
0.5 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 |
1.6 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 |
1.3 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.2 |
1.3 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.2 |
0.8 |
GO:0001923 |
B-1 B cell differentiation(GO:0001923) |
| 0.2 |
0.5 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 0.2 |
2.0 |
GO:0035994 |
response to muscle stretch(GO:0035994) |
| 0.2 |
0.5 |
GO:0007354 |
zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.2 |
0.5 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.2 |
0.9 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 |
1.6 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
0.3 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 |
2.8 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.1 |
1.9 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.1 |
1.4 |
GO:2000381 |
negative regulation of mesoderm development(GO:2000381) |
| 0.1 |
0.8 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
1.0 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.1 |
1.0 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
0.5 |
GO:0035385 |
Roundabout signaling pathway(GO:0035385) |
| 0.1 |
0.5 |
GO:0070245 |
positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 |
0.5 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.1 |
0.9 |
GO:0071910 |
determination of liver left/right asymmetry(GO:0071910) |
| 0.1 |
0.7 |
GO:0097503 |
sialylation(GO:0097503) |
| 0.1 |
0.3 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) |
| 0.1 |
0.4 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.1 |
0.9 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.1 |
0.1 |
GO:0051542 |
elastin biosynthetic process(GO:0051542) |
| 0.1 |
0.6 |
GO:1903347 |
negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 |
0.4 |
GO:0036015 |
response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 |
0.7 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.1 |
0.1 |
GO:1902455 |
negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 |
0.4 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 |
0.7 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 |
0.1 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 |
1.3 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 |
0.8 |
GO:0042473 |
outer ear morphogenesis(GO:0042473) |
| 0.1 |
0.6 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 |
1.1 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.1 |
0.6 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
0.3 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 |
1.1 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.3 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 0.1 |
0.2 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.1 |
2.4 |
GO:0050779 |
RNA destabilization(GO:0050779) |
| 0.1 |
0.5 |
GO:0030916 |
otic vesicle formation(GO:0030916) |
| 0.1 |
1.0 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.1 |
0.3 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 |
4.5 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.1 |
0.4 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.1 |
0.7 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.5 |
GO:0070601 |
centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 |
1.0 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 |
0.4 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 |
0.3 |
GO:1904742 |
regulation of telomeric DNA binding(GO:1904742) |
| 0.1 |
0.1 |
GO:2000812 |
regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 |
1.8 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 |
0.5 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 |
0.3 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.1 |
0.5 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 |
0.3 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 |
0.4 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 |
0.4 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 |
0.8 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 |
0.6 |
GO:0042590 |
antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 |
0.3 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 |
0.3 |
GO:2001045 |
closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 |
0.8 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.5 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 |
0.8 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.1 |
0.3 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 |
1.9 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.1 |
0.6 |
GO:0002591 |
positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.1 |
1.5 |
GO:0031290 |
retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 |
0.1 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 |
1.9 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.1 |
0.2 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 |
0.8 |
GO:0006517 |
protein deglycosylation(GO:0006517) |
| 0.1 |
0.5 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
0.2 |
GO:0006558 |
L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.1 |
0.3 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 |
0.5 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) |
| 0.1 |
0.2 |
GO:0009212 |
dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 |
1.2 |
GO:0071451 |
cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.1 |
0.3 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 |
0.2 |
GO:1904154 |
protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 |
0.4 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 |
1.9 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 |
0.1 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 |
0.1 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) alveolar secondary septum development(GO:0061144) |
| 0.1 |
0.4 |
GO:0048703 |
embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.1 |
0.7 |
GO:1903672 |
positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 |
0.2 |
GO:0045005 |
DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.1 |
0.5 |
GO:0015675 |
nickel cation transport(GO:0015675) |
| 0.1 |
0.4 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 |
0.2 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.1 |
0.3 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.1 |
0.5 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 |
1.3 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
0.8 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.1 |
0.2 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 |
0.3 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
0.3 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.1 |
0.3 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.1 |
0.5 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 |
0.8 |
GO:0044539 |
long-chain fatty acid import(GO:0044539) |
| 0.1 |
1.4 |
GO:1900027 |
regulation of ruffle assembly(GO:1900027) |
| 0.1 |
0.9 |
GO:2000300 |
regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 |
2.3 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 |
0.3 |
GO:0010025 |
wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 |
0.6 |
GO:0033689 |
negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 |
1.6 |
GO:0001709 |
cell fate determination(GO:0001709) |
| 0.1 |
0.9 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 |
0.6 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.1 |
0.3 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 |
0.3 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.1 |
0.5 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 |
0.6 |
GO:0051571 |
positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.1 |
0.5 |
GO:0036123 |
histone H3-K9 dimethylation(GO:0036123) |
| 0.1 |
0.1 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 |
0.1 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.1 |
0.3 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
1.1 |
GO:0001779 |
natural killer cell differentiation(GO:0001779) |
| 0.1 |
0.2 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 |
0.6 |
GO:0072520 |
seminiferous tubule development(GO:0072520) |
| 0.1 |
0.2 |
GO:0060830 |
ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 |
0.2 |
GO:0044829 |
positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 |
0.5 |
GO:0035562 |
negative regulation of chromatin binding(GO:0035562) |
| 0.1 |
1.0 |
GO:0048255 |
mRNA stabilization(GO:0048255) |
| 0.1 |
0.4 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.1 |
2.0 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 |
0.1 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 |
0.4 |
GO:0003413 |
chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.1 |
0.6 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 |
0.2 |
GO:0060479 |
lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.1 |
0.7 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.1 |
0.8 |
GO:0060438 |
trachea development(GO:0060438) |
| 0.1 |
0.5 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.1 |
0.3 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 |
0.4 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 |
0.4 |
GO:0097237 |
cellular response to toxic substance(GO:0097237) |
| 0.0 |
1.6 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.2 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.0 |
0.3 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 |
0.1 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.0 |
0.4 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.2 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.0 |
1.1 |
GO:0044342 |
type B pancreatic cell proliferation(GO:0044342) |
| 0.0 |
0.5 |
GO:0018279 |
protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 |
1.6 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.9 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.0 |
0.1 |
GO:0001787 |
natural killer cell proliferation(GO:0001787) |
| 0.0 |
0.5 |
GO:0002087 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 |
0.1 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.2 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 |
0.6 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.0 |
0.2 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.0 |
0.4 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.3 |
GO:0006699 |
bile acid biosynthetic process(GO:0006699) |
| 0.0 |
0.4 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.4 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 |
3.5 |
GO:0043149 |
contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 |
0.1 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.0 |
0.3 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.0 |
0.5 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.5 |
GO:0060746 |
parental behavior(GO:0060746) |
| 0.0 |
0.4 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 |
0.3 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
1.2 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.1 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.0 |
0.4 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.0 |
0.3 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.1 |
GO:0046950 |
cellular ketone body metabolic process(GO:0046950) |
| 0.0 |
0.6 |
GO:0061036 |
positive regulation of cartilage development(GO:0061036) |
| 0.0 |
0.2 |
GO:0060059 |
embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 |
0.1 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 |
0.7 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.1 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.0 |
1.0 |
GO:0043044 |
ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 |
0.5 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.3 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.1 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 |
0.3 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.1 |
GO:1904395 |
regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 |
0.2 |
GO:0043247 |
protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 |
0.6 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.1 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 |
0.1 |
GO:0003057 |
regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 |
0.4 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.4 |
GO:0001954 |
positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 |
0.3 |
GO:0098868 |
endochondral bone growth(GO:0003416) bone growth(GO:0098868) |
| 0.0 |
0.7 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.0 |
0.6 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.0 |
0.3 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 |
0.4 |
GO:0007080 |
mitotic metaphase plate congression(GO:0007080) |
| 0.0 |
1.0 |
GO:0048663 |
neuron fate commitment(GO:0048663) |
| 0.0 |
0.3 |
GO:0010832 |
negative regulation of myotube differentiation(GO:0010832) |
| 0.0 |
1.1 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.2 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.1 |
GO:0070234 |
positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 |
0.2 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.1 |
GO:0090292 |
nuclear migration along microfilament(GO:0031022) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.1 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.0 |
0.1 |
GO:0042756 |
positive regulation of heat generation(GO:0031652) drinking behavior(GO:0042756) |
| 0.0 |
0.2 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.0 |
0.4 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.0 |
1.1 |
GO:1902850 |
mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 |
0.5 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.1 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
2.0 |
GO:0050885 |
neuromuscular process controlling balance(GO:0050885) |
| 0.0 |
0.1 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 |
0.1 |
GO:2000015 |
regulation of determination of dorsal identity(GO:2000015) |
| 0.0 |
0.4 |
GO:0022617 |
extracellular matrix disassembly(GO:0022617) |
| 0.0 |
0.4 |
GO:0090102 |
cochlea development(GO:0090102) |
| 0.0 |
0.1 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 |
0.5 |
GO:0097009 |
energy homeostasis(GO:0097009) |
| 0.0 |
0.2 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.5 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.0 |
0.2 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.0 |
0.1 |
GO:0033260 |
nuclear DNA replication(GO:0033260) |
| 0.0 |
0.3 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.4 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.7 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.1 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.0 |
0.1 |
GO:0032898 |
neurotrophin production(GO:0032898) nerve growth factor production(GO:0032902) |
| 0.0 |
0.2 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 |
0.1 |
GO:0039703 |
viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 |
1.1 |
GO:0001676 |
long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 |
0.8 |
GO:2000649 |
regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 |
0.2 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.6 |
GO:0043297 |
apical junction assembly(GO:0043297) |
| 0.0 |
0.1 |
GO:0060347 |
heart trabecula formation(GO:0060347) |
| 0.0 |
0.0 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 |
0.2 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.2 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.0 |
0.2 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.1 |
GO:2000269 |
regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 |
1.2 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.6 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 |
0.1 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.0 |
0.2 |
GO:0006515 |
misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 |
0.2 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.0 |
0.4 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.0 |
0.1 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.3 |
GO:1902236 |
negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 |
0.4 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.1 |
GO:1900121 |
regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 |
0.1 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.0 |
0.4 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.4 |
GO:0003170 |
heart valve development(GO:0003170) |
| 0.0 |
0.1 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 0.0 |
0.2 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.1 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 |
0.4 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.0 |
0.1 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
1.2 |
GO:0002230 |
positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 |
0.2 |
GO:1900452 |
regulation of long term synaptic depression(GO:1900452) |
| 0.0 |
0.1 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.4 |
GO:0032784 |
regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 |
0.4 |
GO:1904894 |
positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 |
0.3 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.2 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.0 |
0.1 |
GO:0032402 |
melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 |
0.0 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.0 |
0.0 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 |
0.0 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 |
0.5 |
GO:0006306 |
DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 |
0.0 |
GO:0021830 |
substrate-independent telencephalic tangential migration(GO:0021826) interneuron migration from the subpallium to the cortex(GO:0021830) substrate-independent telencephalic tangential interneuron migration(GO:0021843) |
| 0.0 |
0.5 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
0.0 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 |
0.2 |
GO:0003254 |
regulation of membrane depolarization(GO:0003254) |
| 0.0 |
0.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.3 |
GO:0050729 |
positive regulation of inflammatory response(GO:0050729) |
| 0.0 |
1.1 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.6 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |