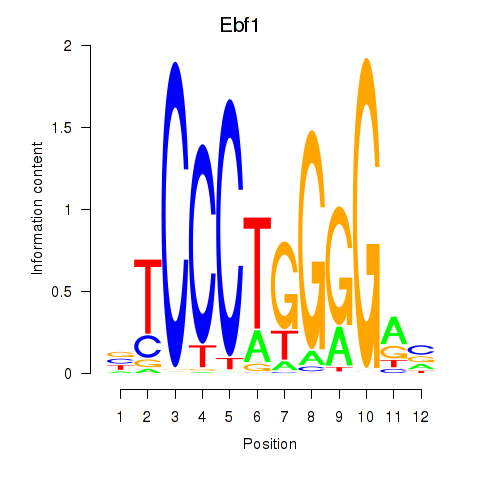
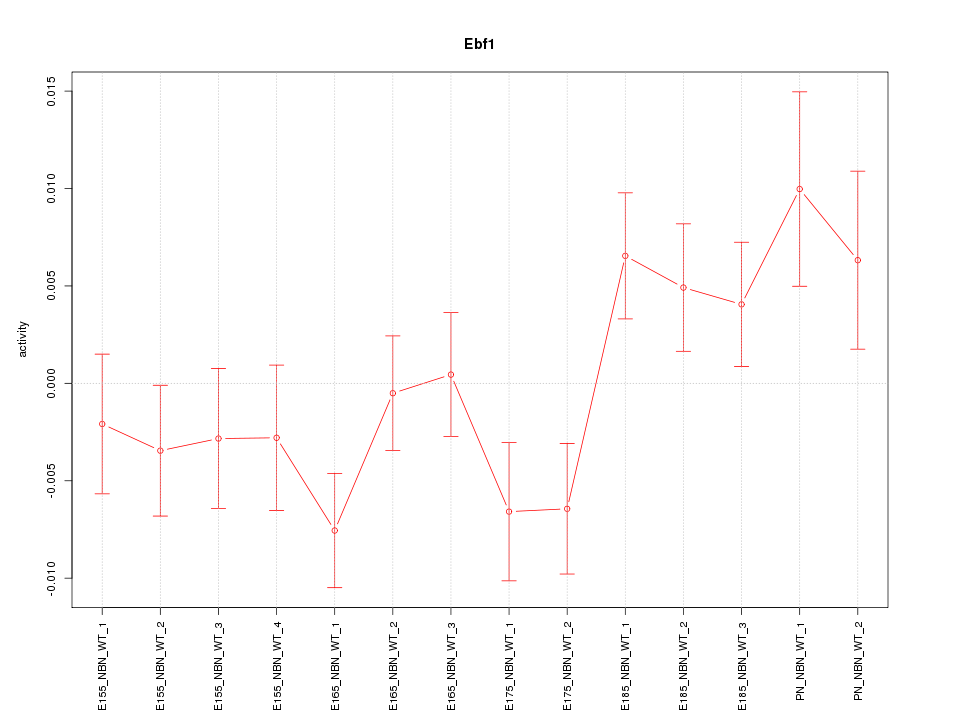
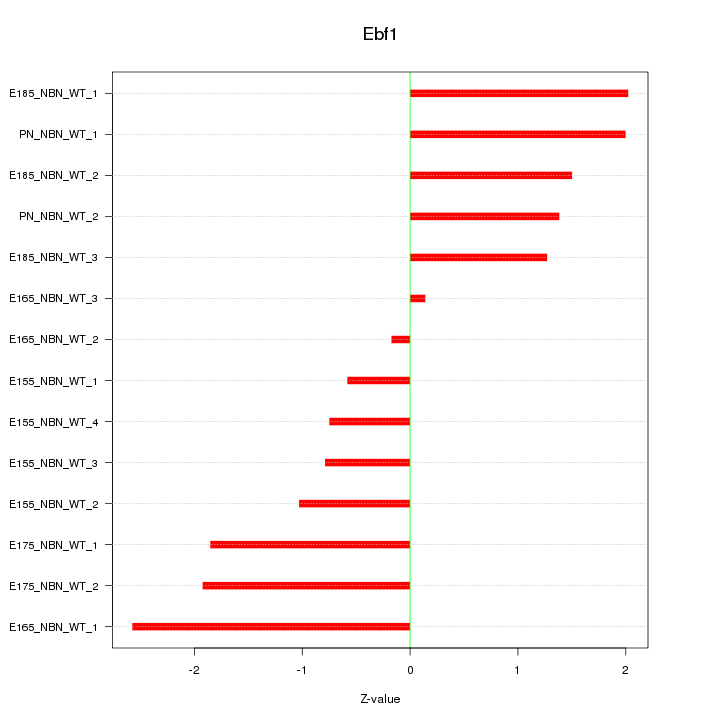
| 1.3 |
6.7 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 1.2 |
3.5 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.0 |
2.9 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.8 |
3.1 |
GO:0038145 |
macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.7 |
2.9 |
GO:2000525 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.7 |
2.1 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.6 |
1.9 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.6 |
0.6 |
GO:0060032 |
smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) notochord regression(GO:0060032) |
| 0.6 |
3.3 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.5 |
1.0 |
GO:2000157 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.5 |
1.5 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.5 |
0.5 |
GO:0032829 |
regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.5 |
1.4 |
GO:1900158 |
negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.5 |
0.9 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.5 |
0.9 |
GO:0035771 |
interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.5 |
1.4 |
GO:0072180 |
mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) |
| 0.5 |
1.8 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.4 |
3.1 |
GO:0042590 |
antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.4 |
1.8 |
GO:0071280 |
cellular response to copper ion(GO:0071280) |
| 0.4 |
2.2 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.4 |
1.2 |
GO:0010533 |
regulation of activation of Janus kinase activity(GO:0010533) regulation of activation of JAK2 kinase activity(GO:0010534) |
| 0.4 |
2.4 |
GO:0072224 |
metanephric glomerulus development(GO:0072224) |
| 0.4 |
1.6 |
GO:1900426 |
positive regulation of defense response to bacterium(GO:1900426) |
| 0.4 |
1.9 |
GO:0046864 |
retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.4 |
1.5 |
GO:0061626 |
pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.4 |
1.5 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.4 |
1.9 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.4 |
1.8 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.4 |
1.1 |
GO:0006001 |
fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.4 |
1.8 |
GO:0021905 |
pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) |
| 0.4 |
0.4 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.3 |
2.4 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.3 |
1.4 |
GO:0071449 |
cellular response to lipid hydroperoxide(GO:0071449) |
| 0.3 |
1.0 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.3 |
1.0 |
GO:0052428 |
modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.3 |
1.3 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 |
2.2 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.3 |
1.9 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.3 |
0.9 |
GO:0016102 |
retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.3 |
0.9 |
GO:0036118 |
hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.3 |
0.9 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 |
1.2 |
GO:0003430 |
tolerance induction to self antigen(GO:0002513) growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.3 |
1.7 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.3 |
0.9 |
GO:1902037 |
negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.3 |
1.7 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.3 |
2.8 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.3 |
0.3 |
GO:0002488 |
antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) |
| 0.3 |
0.8 |
GO:0002606 |
positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.3 |
1.1 |
GO:0048143 |
astrocyte activation(GO:0048143) |
| 0.3 |
2.7 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.3 |
0.8 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 |
0.5 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.3 |
0.8 |
GO:0019043 |
establishment of viral latency(GO:0019043) |
| 0.3 |
1.1 |
GO:0010700 |
negative regulation of norepinephrine secretion(GO:0010700) |
| 0.3 |
1.3 |
GO:1903012 |
positive regulation of bone development(GO:1903012) |
| 0.3 |
1.0 |
GO:0048819 |
regulation of hair follicle maturation(GO:0048819) regulation of catagen(GO:0051794) |
| 0.3 |
1.0 |
GO:0071680 |
response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.2 |
1.0 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 |
0.7 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.2 |
0.7 |
GO:0046544 |
regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) development of secondary male sexual characteristics(GO:0046544) |
| 0.2 |
0.5 |
GO:0035880 |
embryonic nail plate morphogenesis(GO:0035880) |
| 0.2 |
0.2 |
GO:0001787 |
natural killer cell proliferation(GO:0001787) |
| 0.2 |
0.7 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.2 |
0.7 |
GO:0010989 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.2 |
0.7 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.2 |
1.2 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.2 |
1.1 |
GO:0060355 |
positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.2 |
1.6 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.2 |
0.7 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.2 |
0.9 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.2 |
0.9 |
GO:2000504 |
positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 |
0.6 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.2 |
0.6 |
GO:0032240 |
mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 |
1.9 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.2 |
0.6 |
GO:0071603 |
endothelial cell-cell adhesion(GO:0071603) |
| 0.2 |
0.6 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.2 |
3.7 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.2 |
0.4 |
GO:0000720 |
pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.2 |
0.6 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.2 |
0.8 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.2 |
0.9 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.2 |
0.6 |
GO:0010898 |
positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 |
0.8 |
GO:0060051 |
negative regulation of protein glycosylation(GO:0060051) |
| 0.2 |
0.8 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.2 |
0.6 |
GO:0060753 |
regulation of mast cell chemotaxis(GO:0060753) |
| 0.2 |
0.7 |
GO:1904996 |
positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.2 |
1.8 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 |
0.9 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.2 |
0.5 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.2 |
1.3 |
GO:0002484 |
antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.2 |
0.7 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 |
0.3 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 |
0.7 |
GO:0032901 |
positive regulation of neurotrophin production(GO:0032901) |
| 0.2 |
0.9 |
GO:0006167 |
AMP biosynthetic process(GO:0006167) |
| 0.2 |
0.5 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.2 |
0.5 |
GO:0060160 |
negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.2 |
1.0 |
GO:1901979 |
regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.2 |
0.2 |
GO:0045014 |
carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 |
0.2 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.2 |
0.3 |
GO:0061043 |
regulation of vascular wound healing(GO:0061043) |
| 0.2 |
0.5 |
GO:0032962 |
positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.2 |
0.5 |
GO:0060785 |
regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.2 |
1.0 |
GO:0061304 |
retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 |
0.8 |
GO:0071499 |
response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.2 |
0.6 |
GO:1904426 |
positive regulation of GTP binding(GO:1904426) regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.2 |
0.5 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 |
0.5 |
GO:0051006 |
positive regulation of lipoprotein lipase activity(GO:0051006) |
| 0.2 |
0.5 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.2 |
2.7 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.2 |
0.6 |
GO:0010593 |
negative regulation of lamellipodium assembly(GO:0010593) |
| 0.2 |
0.6 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.7 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.1 |
0.4 |
GO:0033603 |
positive regulation of dopamine secretion(GO:0033603) |
| 0.1 |
1.0 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 |
0.1 |
GO:0010039 |
response to iron ion(GO:0010039) |
| 0.1 |
0.4 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.1 |
0.4 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 |
0.3 |
GO:0042891 |
antibiotic transport(GO:0042891) |
| 0.1 |
1.4 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 |
0.1 |
GO:0007161 |
calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 |
0.6 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.1 |
1.7 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.1 |
0.7 |
GO:0071394 |
cellular response to testosterone stimulus(GO:0071394) |
| 0.1 |
1.0 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.1 |
1.3 |
GO:0043206 |
extracellular fibril organization(GO:0043206) |
| 0.1 |
0.5 |
GO:0007262 |
STAT protein import into nucleus(GO:0007262) |
| 0.1 |
0.4 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 |
0.5 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.1 |
0.3 |
GO:0015920 |
lipopolysaccharide transport(GO:0015920) |
| 0.1 |
0.3 |
GO:0003433 |
chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 |
1.7 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 |
0.8 |
GO:1902669 |
positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.1 |
0.4 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 |
0.6 |
GO:0032911 |
negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.6 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.1 |
0.4 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 |
0.2 |
GO:0060535 |
trachea cartilage morphogenesis(GO:0060535) |
| 0.1 |
1.2 |
GO:0042416 |
dopamine biosynthetic process(GO:0042416) |
| 0.1 |
0.4 |
GO:0033602 |
negative regulation of dopamine secretion(GO:0033602) |
| 0.1 |
0.4 |
GO:0002924 |
negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.1 |
0.5 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 |
0.6 |
GO:0034380 |
high-density lipoprotein particle assembly(GO:0034380) |
| 0.1 |
0.6 |
GO:0034123 |
positive regulation of toll-like receptor signaling pathway(GO:0034123) |
| 0.1 |
0.6 |
GO:0032237 |
activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 |
0.5 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 |
0.2 |
GO:0046121 |
deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 |
0.4 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 |
0.2 |
GO:0031394 |
positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 |
0.2 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 |
0.5 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
0.6 |
GO:0014050 |
negative regulation of glutamate secretion(GO:0014050) |
| 0.1 |
0.3 |
GO:0046104 |
thymidine metabolic process(GO:0046104) |
| 0.1 |
3.2 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 |
0.7 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 |
0.2 |
GO:0044034 |
negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 |
0.4 |
GO:0007039 |
protein catabolic process in the vacuole(GO:0007039) |
| 0.1 |
0.6 |
GO:0019370 |
leukotriene biosynthetic process(GO:0019370) |
| 0.1 |
1.1 |
GO:0043312 |
neutrophil degranulation(GO:0043312) |
| 0.1 |
1.4 |
GO:0042346 |
positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.1 |
0.7 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.1 |
0.3 |
GO:0051342 |
regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 |
0.6 |
GO:0003065 |
positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 |
0.5 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.1 |
0.2 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 |
0.3 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 |
0.3 |
GO:0070476 |
rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 |
0.9 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.1 |
0.1 |
GO:0002517 |
T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) |
| 0.1 |
0.6 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.1 |
0.4 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.1 |
0.6 |
GO:0098838 |
reduced folate transmembrane transport(GO:0098838) |
| 0.1 |
0.4 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.1 |
0.1 |
GO:1901228 |
positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 |
0.5 |
GO:0042758 |
long-chain fatty acid catabolic process(GO:0042758) |
| 0.1 |
0.7 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.1 |
0.4 |
GO:1903121 |
regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) |
| 0.1 |
1.5 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 |
0.9 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.1 |
0.6 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.1 |
1.0 |
GO:0060033 |
anatomical structure regression(GO:0060033) |
| 0.1 |
0.5 |
GO:0060346 |
bone trabecula formation(GO:0060346) |
| 0.1 |
0.2 |
GO:0033088 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 |
0.2 |
GO:0006702 |
androgen biosynthetic process(GO:0006702) |
| 0.1 |
0.7 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
0.4 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 |
0.3 |
GO:0010991 |
regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
0.3 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.1 |
0.7 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 |
1.6 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.1 |
0.6 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
0.3 |
GO:0060406 |
positive regulation of penile erection(GO:0060406) |
| 0.1 |
0.4 |
GO:0048711 |
positive regulation of astrocyte differentiation(GO:0048711) |
| 0.1 |
0.7 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 |
0.9 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.1 |
0.4 |
GO:0071316 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) cellular response to nicotine(GO:0071316) |
| 0.1 |
0.3 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.1 |
0.9 |
GO:0002716 |
negative regulation of natural killer cell mediated immunity(GO:0002716) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.1 |
0.9 |
GO:0060294 |
cilium movement involved in cell motility(GO:0060294) |
| 0.1 |
1.6 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 |
1.1 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.1 |
0.5 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 |
0.4 |
GO:0051771 |
negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 |
0.5 |
GO:0070447 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 |
0.6 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.1 |
0.3 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 0.1 |
0.3 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.1 |
1.2 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.1 |
0.2 |
GO:1902956 |
regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.1 |
0.5 |
GO:0006046 |
N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 |
2.2 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 |
0.3 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.1 |
0.4 |
GO:0009068 |
aspartate family amino acid catabolic process(GO:0009068) |
| 0.1 |
0.3 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 |
0.2 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 |
0.2 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 |
0.3 |
GO:0046469 |
platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.1 |
0.2 |
GO:0015675 |
nickel cation transport(GO:0015675) |
| 0.1 |
0.3 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.1 |
0.6 |
GO:0030174 |
regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 |
0.4 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
1.7 |
GO:0021516 |
dorsal spinal cord development(GO:0021516) |
| 0.1 |
0.6 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 |
0.2 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
2.1 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 |
0.2 |
GO:0031038 |
myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 |
0.5 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.1 |
0.2 |
GO:0035696 |
monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.1 |
0.2 |
GO:0090272 |
negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.1 |
0.2 |
GO:0035106 |
operant conditioning(GO:0035106) |
| 0.1 |
0.1 |
GO:0061642 |
chemoattraction of axon(GO:0061642) |
| 0.1 |
0.7 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
0.4 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.1 |
0.1 |
GO:2001274 |
negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 |
0.3 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 |
0.6 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 |
0.2 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 |
0.3 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.1 |
0.4 |
GO:0021936 |
regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.1 |
0.4 |
GO:0045577 |
regulation of B cell differentiation(GO:0045577) |
| 0.1 |
0.7 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.1 |
0.4 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 |
0.2 |
GO:0042196 |
dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.1 |
0.7 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 |
0.3 |
GO:0042985 |
negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 |
0.1 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 |
0.1 |
GO:0097460 |
ferrous iron import(GO:0070627) ferrous iron import into cell(GO:0097460) |
| 0.1 |
0.3 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 |
0.2 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 |
0.4 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 |
0.2 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 |
0.2 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 |
0.7 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.1 |
0.1 |
GO:0002922 |
positive regulation of humoral immune response(GO:0002922) |
| 0.1 |
0.2 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.1 |
0.4 |
GO:0002347 |
response to tumor cell(GO:0002347) |
| 0.1 |
0.2 |
GO:0035360 |
positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 |
0.2 |
GO:0006691 |
leukotriene metabolic process(GO:0006691) |
| 0.1 |
0.2 |
GO:1990481 |
mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 |
0.5 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 |
0.3 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.1 |
0.3 |
GO:0006528 |
asparagine metabolic process(GO:0006528) |
| 0.1 |
0.3 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 |
0.1 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.1 |
1.2 |
GO:0042345 |
regulation of NF-kappaB import into nucleus(GO:0042345) NF-kappaB import into nucleus(GO:0042348) |
| 0.1 |
0.2 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.1 |
1.0 |
GO:0001913 |
T cell mediated cytotoxicity(GO:0001913) |
| 0.1 |
0.6 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 |
0.3 |
GO:1900186 |
negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.1 |
0.4 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 |
1.0 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.1 |
0.2 |
GO:0044650 |
virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 |
0.2 |
GO:0021514 |
ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 |
0.3 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 |
0.1 |
GO:0035793 |
regulation of phosphatidylinositol biosynthetic process(GO:0010511) negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.1 |
0.4 |
GO:0071493 |
cellular response to UV-B(GO:0071493) |
| 0.1 |
0.1 |
GO:0033484 |
nitric oxide homeostasis(GO:0033484) |
| 0.1 |
0.1 |
GO:0032898 |
neurotrophin production(GO:0032898) |
| 0.1 |
0.4 |
GO:0014048 |
regulation of glutamate secretion(GO:0014048) |
| 0.1 |
0.1 |
GO:2001187 |
positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.1 |
0.5 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 |
0.5 |
GO:0061298 |
retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 |
3.7 |
GO:0019882 |
antigen processing and presentation(GO:0019882) |
| 0.1 |
0.4 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.1 |
0.2 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.1 |
0.2 |
GO:0030049 |
muscle filament sliding(GO:0030049) |
| 0.1 |
0.5 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.1 |
0.2 |
GO:0019471 |
4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 |
0.5 |
GO:0046060 |
dATP metabolic process(GO:0046060) |
| 0.1 |
0.2 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.1 |
0.7 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 |
0.2 |
GO:0044331 |
cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.1 |
0.2 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.1 |
0.1 |
GO:0016114 |
terpenoid biosynthetic process(GO:0016114) |
| 0.1 |
0.2 |
GO:0010519 |
negative regulation of phospholipase activity(GO:0010519) |
| 0.1 |
0.2 |
GO:1902047 |
polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 |
0.4 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 |
0.3 |
GO:0042510 |
tyrosine phosphorylation of Stat1 protein(GO:0042508) regulation of tyrosine phosphorylation of Stat1 protein(GO:0042510) |
| 0.1 |
0.3 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.5 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.1 |
0.1 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.1 |
0.3 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 |
0.4 |
GO:1904707 |
positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 |
0.3 |
GO:0044336 |
canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 |
0.1 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.1 |
0.2 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 |
0.4 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.1 |
0.3 |
GO:0035188 |
blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 |
0.3 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 |
0.2 |
GO:0018158 |
protein oxidation(GO:0018158) |
| 0.1 |
0.1 |
GO:0072061 |
inner medullary collecting duct development(GO:0072061) |
| 0.1 |
0.1 |
GO:0019249 |
lactate metabolic process(GO:0006089) lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.1 |
0.1 |
GO:0060330 |
regulation of response to interferon-gamma(GO:0060330) |
| 0.0 |
1.6 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.7 |
GO:1901186 |
positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 |
0.5 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.0 |
0.1 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.0 |
0.1 |
GO:0021898 |
commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 |
0.2 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.0 |
0.0 |
GO:0036462 |
TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 |
0.5 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 |
0.2 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 |
0.9 |
GO:0045838 |
positive regulation of membrane potential(GO:0045838) |
| 0.0 |
0.3 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 |
0.4 |
GO:0060768 |
regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) |
| 0.0 |
0.7 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 |
0.2 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.0 |
2.1 |
GO:0009142 |
nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 |
0.1 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.0 |
0.1 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 |
0.2 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 |
0.1 |
GO:0072338 |
allantoin metabolic process(GO:0000255) activation of JAK2 kinase activity(GO:0042977) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 |
0.2 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.0 |
0.1 |
GO:2000664 |
positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 |
1.0 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.2 |
GO:0003420 |
regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) |
| 0.0 |
0.1 |
GO:0010725 |
regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.0 |
0.0 |
GO:0070309 |
lens fiber cell morphogenesis(GO:0070309) |
| 0.0 |
0.3 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 |
0.1 |
GO:0032703 |
negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 |
0.4 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 |
0.2 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.2 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 |
0.3 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.0 |
0.4 |
GO:0006560 |
proline metabolic process(GO:0006560) |
| 0.0 |
0.3 |
GO:2001223 |
negative regulation of neuron migration(GO:2001223) |
| 0.0 |
0.2 |
GO:2001015 |
negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 |
0.7 |
GO:0042789 |
mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 |
0.2 |
GO:0090666 |
telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.3 |
GO:0051127 |
regulation of actin nucleation(GO:0051125) positive regulation of actin nucleation(GO:0051127) |
| 0.0 |
0.3 |
GO:2000194 |
regulation of female gonad development(GO:2000194) |
| 0.0 |
0.4 |
GO:0036159 |
inner dynein arm assembly(GO:0036159) |
| 0.0 |
0.3 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.0 |
0.1 |
GO:0042560 |
10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 |
0.1 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 |
0.1 |
GO:0015868 |
purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.1 |
GO:0070375 |
ERK5 cascade(GO:0070375) |
| 0.0 |
2.0 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.3 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 |
1.5 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 |
0.2 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.4 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 |
0.2 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.0 |
0.3 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) |
| 0.0 |
0.4 |
GO:0044406 |
adhesion of symbiont to host(GO:0044406) |
| 0.0 |
0.2 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.0 |
0.4 |
GO:2000505 |
regulation of energy homeostasis(GO:2000505) |
| 0.0 |
0.1 |
GO:0033030 |
negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 |
1.0 |
GO:0035518 |
histone H2A monoubiquitination(GO:0035518) |
| 0.0 |
0.2 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 |
0.2 |
GO:0010801 |
negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 |
0.2 |
GO:0032511 |
late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 |
0.1 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 |
0.2 |
GO:0051096 |
regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 |
0.1 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.0 |
0.2 |
GO:0019346 |
homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 |
0.1 |
GO:0046533 |
regulation of photoreceptor cell differentiation(GO:0046532) negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.0 |
0.2 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.1 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 |
0.1 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 |
0.1 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 |
0.3 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.1 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 |
0.1 |
GO:0060591 |
chondroblast differentiation(GO:0060591) |
| 0.0 |
0.1 |
GO:0045187 |
regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.2 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.3 |
GO:2000678 |
negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 |
0.2 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 |
0.0 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 |
0.1 |
GO:0070245 |
positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 |
0.5 |
GO:0031065 |
positive regulation of histone deacetylation(GO:0031065) |
| 0.0 |
0.1 |
GO:1900017 |
positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 |
0.2 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.3 |
GO:0051307 |
meiotic chromosome separation(GO:0051307) |
| 0.0 |
0.5 |
GO:0001945 |
lymph vessel development(GO:0001945) |
| 0.0 |
0.1 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 |
0.4 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.0 |
0.7 |
GO:0034340 |
response to type I interferon(GO:0034340) |
| 0.0 |
0.2 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.0 |
0.5 |
GO:0035116 |
embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 |
0.6 |
GO:0032287 |
peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 |
0.2 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.2 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 |
0.6 |
GO:0003229 |
ventricular cardiac muscle tissue development(GO:0003229) |
| 0.0 |
0.1 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 |
0.7 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.1 |
GO:0032815 |
negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 |
0.1 |
GO:1901386 |
negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 |
0.3 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.1 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.0 |
0.4 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 |
0.2 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.0 |
0.2 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 |
0.5 |
GO:1901984 |
negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 |
0.1 |
GO:0045624 |
positive regulation of T-helper cell differentiation(GO:0045624) |
| 0.0 |
0.2 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.4 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 |
0.2 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.0 |
0.5 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.2 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.2 |
GO:0042832 |
defense response to protozoan(GO:0042832) |
| 0.0 |
0.1 |
GO:0019230 |
proprioception(GO:0019230) |
| 0.0 |
0.1 |
GO:0036509 |
trimming of terminal mannose on B branch(GO:0036509) |
| 0.0 |
0.1 |
GO:0035166 |
post-embryonic hemopoiesis(GO:0035166) |
| 0.0 |
0.2 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.0 |
0.2 |
GO:0036066 |
protein O-linked fucosylation(GO:0036066) |
| 0.0 |
0.2 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) |
| 0.0 |
0.1 |
GO:0070836 |
caveola assembly(GO:0070836) |
| 0.0 |
0.1 |
GO:0048011 |
neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 |
0.7 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.1 |
GO:0046498 |
S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 |
0.1 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.0 |
0.3 |
GO:0044342 |
type B pancreatic cell proliferation(GO:0044342) |
| 0.0 |
0.1 |
GO:0098661 |
inorganic anion transmembrane transport(GO:0098661) |
| 0.0 |
0.1 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.0 |
0.0 |
GO:0014858 |
positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 |
0.3 |
GO:0034219 |
carbohydrate transmembrane transport(GO:0034219) |
| 0.0 |
0.2 |
GO:0043970 |
histone H3-K9 acetylation(GO:0043970) |
| 0.0 |
0.0 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.0 |
0.2 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.1 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 |
0.1 |
GO:0000393 |
spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 |
0.2 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 |
0.1 |
GO:0061009 |
common bile duct development(GO:0061009) |
| 0.0 |
0.0 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) positive regulation of follicle-stimulating hormone secretion(GO:0046881) follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 |
0.2 |
GO:0007042 |
lysosomal lumen acidification(GO:0007042) |
| 0.0 |
0.1 |
GO:0045655 |
regulation of monocyte differentiation(GO:0045655) |
| 0.0 |
0.2 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.0 |
0.2 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.1 |
GO:0060297 |
regulation of sarcomere organization(GO:0060297) |
| 0.0 |
0.1 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.1 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 0.0 |
0.0 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 |
0.8 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.4 |
GO:0055012 |
ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 |
0.1 |
GO:0051798 |
positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 |
0.2 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.2 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.1 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.0 |
0.1 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 |
0.5 |
GO:0034315 |
regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 |
0.2 |
GO:0045123 |
cellular extravasation(GO:0045123) |
| 0.0 |
0.1 |
GO:0006451 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.2 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.5 |
GO:0060445 |
branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 |
0.2 |
GO:0002082 |
regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 |
0.2 |
GO:1903423 |
positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 |
0.1 |
GO:0035813 |
renal sodium excretion(GO:0035812) regulation of renal sodium excretion(GO:0035813) |
| 0.0 |
0.3 |
GO:0055070 |
copper ion homeostasis(GO:0055070) |
| 0.0 |
0.2 |
GO:0010839 |
negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 |
0.1 |
GO:0035729 |
response to hepatocyte growth factor(GO:0035728) cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 |
0.1 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
0.2 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.6 |
GO:0035456 |
response to interferon-beta(GO:0035456) |
| 0.0 |
0.1 |
GO:0009838 |
abscission(GO:0009838) |
| 0.0 |
0.3 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 |
0.1 |
GO:0048745 |
smooth muscle tissue development(GO:0048745) |
| 0.0 |
0.1 |
GO:0032682 |
negative regulation of chemokine production(GO:0032682) |
| 0.0 |
0.1 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 |
0.1 |
GO:0033605 |
positive regulation of catecholamine secretion(GO:0033605) |
| 0.0 |
0.4 |
GO:0097284 |
hepatocyte apoptotic process(GO:0097284) |
| 0.0 |
0.4 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.2 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.0 |
0.1 |
GO:1903755 |
regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.0 |
0.3 |
GO:0033628 |
regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.0 |
0.4 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.1 |
GO:0036500 |
ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 |
0.2 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.0 |
0.1 |
GO:0097503 |
sialylation(GO:0097503) |
| 0.0 |
0.2 |
GO:0009299 |
mRNA transcription(GO:0009299) |
| 0.0 |
0.2 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 |
0.2 |
GO:0060441 |
epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 |
0.1 |
GO:1904948 |
midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 |
0.1 |
GO:0010804 |
negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 |
0.2 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.2 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.0 |
0.5 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 |
0.1 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.0 |
0.2 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.0 |
0.0 |
GO:0051956 |
negative regulation of amino acid transport(GO:0051956) |
| 0.0 |
0.1 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.0 |
0.2 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.0 |
0.1 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 |
0.6 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.2 |
GO:0036093 |
germ cell proliferation(GO:0036093) |
| 0.0 |
0.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.3 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 |
0.1 |
GO:0045900 |
negative regulation of translational elongation(GO:0045900) |
| 0.0 |
0.1 |
GO:0060821 |
inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.0 |
0.3 |
GO:1902042 |
negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 |
0.1 |
GO:0045687 |
positive regulation of glial cell differentiation(GO:0045687) |
| 0.0 |
0.2 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.0 |
0.0 |
GO:0097494 |
regulation of vesicle size(GO:0097494) |
| 0.0 |
0.2 |
GO:0042246 |
tissue regeneration(GO:0042246) |
| 0.0 |
0.3 |
GO:0007080 |
mitotic metaphase plate congression(GO:0007080) |
| 0.0 |
0.2 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.0 |
0.1 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 |
0.3 |
GO:0046470 |
phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 |
0.1 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.0 |
0.2 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
0.4 |
GO:0002053 |
positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 |
0.1 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 |
0.5 |
GO:1902186 |
regulation of viral release from host cell(GO:1902186) |
| 0.0 |
0.1 |
GO:2000650 |
negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 |
0.1 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 |
0.2 |
GO:0050434 |
positive regulation of viral transcription(GO:0050434) |
| 0.0 |
0.4 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.9 |
GO:0006635 |
fatty acid beta-oxidation(GO:0006635) |
| 0.0 |
0.0 |
GO:0006549 |
isoleucine metabolic process(GO:0006549) |
| 0.0 |
0.1 |
GO:0006627 |
protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 |
0.8 |
GO:0071230 |
cellular response to amino acid stimulus(GO:0071230) |
| 0.0 |
0.1 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.1 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 |
0.0 |
GO:0035584 |
calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 |
0.2 |
GO:0002063 |
chondrocyte development(GO:0002063) |
| 0.0 |
0.1 |
GO:2001135 |
regulation of endocytic recycling(GO:2001135) |
| 0.0 |
0.1 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
1.1 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.2 |
GO:2000772 |
regulation of cellular senescence(GO:2000772) |
| 0.0 |
0.4 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.1 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.1 |
GO:0070278 |
extracellular matrix constituent secretion(GO:0070278) |
| 0.0 |
0.0 |
GO:0070307 |
lens fiber cell development(GO:0070307) |
| 0.0 |
0.2 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.1 |
GO:0022615 |
protein to membrane docking(GO:0022615) |
| 0.0 |
0.2 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.2 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.0 |
0.5 |
GO:0032755 |
positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 |
0.1 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.1 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 |
0.2 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.1 |
GO:1903975 |
regulation of glial cell migration(GO:1903975) |
| 0.0 |
0.1 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.0 |
0.0 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 |
0.1 |
GO:0031055 |
chromatin remodeling at centromere(GO:0031055) |
| 0.0 |
0.1 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 |
0.0 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 |
0.1 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 0.0 |
0.2 |
GO:0050919 |
negative chemotaxis(GO:0050919) |
| 0.0 |
0.2 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.2 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.4 |
GO:0060479 |
lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.0 |
0.0 |
GO:0006000 |
fructose metabolic process(GO:0006000) |
| 0.0 |
0.3 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.2 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.1 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.1 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.1 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
0.1 |
GO:0030813 |
positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of nucleoside metabolic process(GO:0045979) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) positive regulation of ATP metabolic process(GO:1903580) |
| 0.0 |
0.0 |
GO:0042359 |
vitamin D metabolic process(GO:0042359) |
| 0.0 |
0.3 |
GO:0022904 |
respiratory electron transport chain(GO:0022904) |
| 0.0 |
0.0 |
GO:0051541 |
elastin metabolic process(GO:0051541) |
| 0.0 |
0.1 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.0 |
0.2 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.3 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.3 |
GO:0015909 |
long-chain fatty acid transport(GO:0015909) |
| 0.0 |
0.1 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.1 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 |
0.4 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
0.1 |
GO:0006537 |
glutamate biosynthetic process(GO:0006537) |
| 0.0 |
0.1 |
GO:0042538 |
hyperosmotic salinity response(GO:0042538) |
| 0.0 |
0.1 |
GO:0051788 |
response to misfolded protein(GO:0051788) |
| 0.0 |
0.1 |
GO:0035767 |
endothelial cell chemotaxis(GO:0035767) |
| 0.0 |
0.1 |
GO:2000300 |
regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 |
0.0 |
GO:0006097 |
glyoxylate cycle(GO:0006097) glyoxylate metabolic process(GO:0046487) |
| 0.0 |
0.1 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 |
0.1 |
GO:0006636 |
unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 |
0.1 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.1 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.0 |
0.0 |
GO:0090467 |
L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 |
0.2 |
GO:0071470 |
cellular response to osmotic stress(GO:0071470) |
| 0.0 |
0.1 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.1 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.1 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.1 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 |
0.1 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
0.1 |
GO:0070200 |
establishment of protein localization to telomere(GO:0070200) |
| 0.0 |
0.2 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.1 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 |
0.1 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.1 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.1 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
0.1 |
GO:2000381 |
negative regulation of mesoderm development(GO:2000381) |
| 0.0 |
0.1 |
GO:0032464 |
regulation of protein homooligomerization(GO:0032462) positive regulation of protein homooligomerization(GO:0032464) |