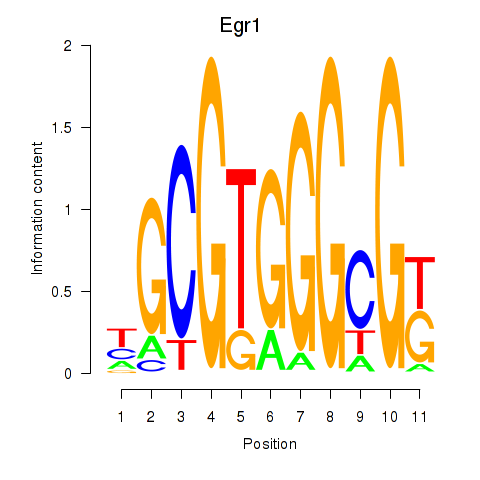
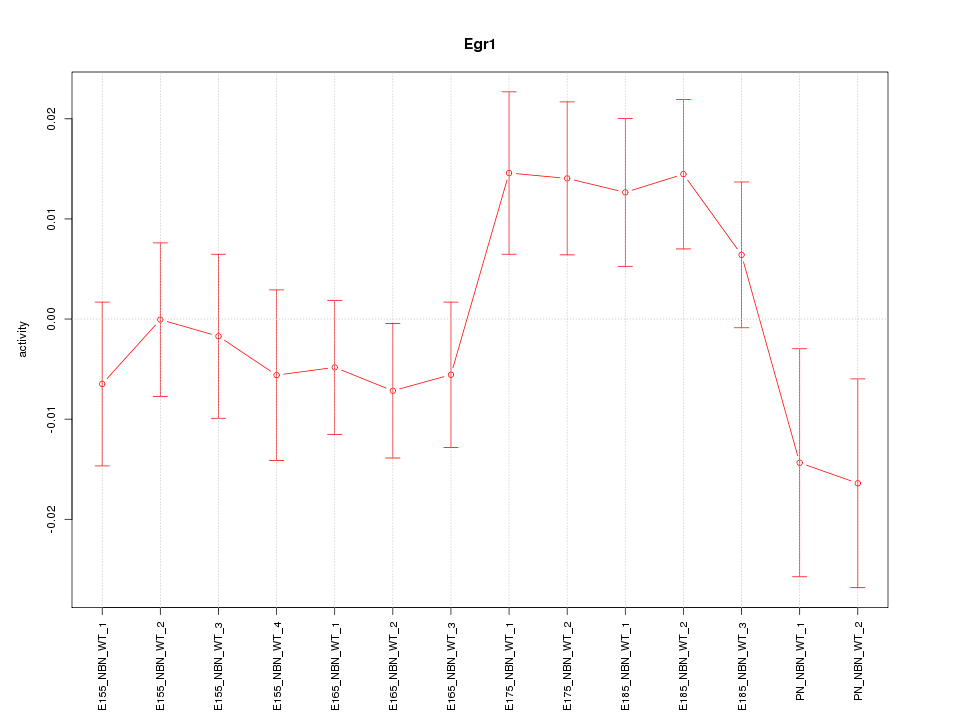
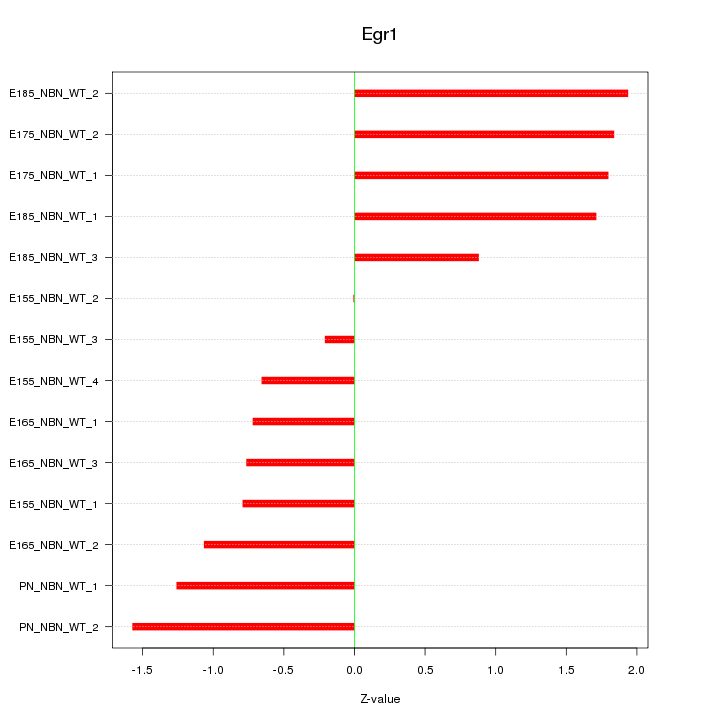
| 0.7 |
2.0 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.5 |
1.9 |
GO:2000313 |
fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.4 |
1.2 |
GO:1900039 |
positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.3 |
0.3 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.3 |
1.4 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.3 |
0.8 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.3 |
1.1 |
GO:0010040 |
response to iron(II) ion(GO:0010040) positive regulation of hydrogen peroxide metabolic process(GO:0010726) negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) cellular response to copper ion(GO:0071280) regulation of peroxidase activity(GO:2000468) |
| 0.2 |
1.2 |
GO:2000325 |
regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 |
2.2 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 |
0.6 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.2 |
1.0 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 |
1.2 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.2 |
1.0 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 |
0.9 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 |
0.9 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.2 |
0.5 |
GO:0033159 |
negative regulation of protein import into nucleus, translocation(GO:0033159) protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 |
0.5 |
GO:0097151 |
positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.2 |
0.7 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 |
0.5 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.2 |
0.7 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.2 |
1.9 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.9 |
GO:1900451 |
positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 |
0.4 |
GO:0010512 |
negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.1 |
0.6 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 |
0.7 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.1 |
0.4 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 |
0.4 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 |
0.8 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.4 |
GO:0098528 |
skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 |
0.5 |
GO:0090365 |
regulation of mRNA modification(GO:0090365) |
| 0.1 |
1.1 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
0.9 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.1 |
2.4 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 |
0.8 |
GO:0032796 |
uropod organization(GO:0032796) |
| 0.1 |
0.6 |
GO:2001025 |
positive regulation of response to drug(GO:2001025) |
| 0.1 |
0.5 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.1 |
0.8 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.1 |
1.0 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.1 |
0.9 |
GO:0060732 |
positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 |
2.1 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 |
0.6 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.1 |
0.4 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 |
1.1 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.1 |
0.5 |
GO:0072368 |
regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.1 |
0.6 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.1 |
0.3 |
GO:0045014 |
ectoderm formation(GO:0001705) detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.1 |
0.8 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.1 |
0.6 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
0.4 |
GO:0070086 |
ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 |
0.4 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 |
0.3 |
GO:1902109 |
negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 |
2.2 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.1 |
0.4 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
0.5 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 |
1.3 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 |
0.2 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.1 |
0.2 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.1 |
0.3 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 |
1.6 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.1 |
0.4 |
GO:0032532 |
regulation of microvillus length(GO:0032532) |
| 0.1 |
0.5 |
GO:1900116 |
sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 |
0.4 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 |
0.4 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.1 |
0.2 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 |
0.2 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 |
0.5 |
GO:0097369 |
sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
0.8 |
GO:0032688 |
negative regulation of interferon-beta production(GO:0032688) |
| 0.1 |
0.5 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.2 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 |
0.6 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.1 |
0.1 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) |
| 0.1 |
0.2 |
GO:1903966 |
monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 |
0.9 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 |
0.7 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
0.7 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.1 |
0.3 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.1 |
0.7 |
GO:0032525 |
somite rostral/caudal axis specification(GO:0032525) |
| 0.1 |
0.1 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 |
0.4 |
GO:0001956 |
positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.1 |
0.1 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 |
0.5 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.2 |
GO:1900673 |
pons maturation(GO:0021586) olefin metabolic process(GO:1900673) |
| 0.1 |
0.5 |
GO:0033008 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 |
0.6 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.1 |
0.2 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 |
0.2 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 |
0.4 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.1 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.0 |
0.8 |
GO:1900273 |
positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 |
0.2 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.6 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.3 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.0 |
0.1 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.0 |
0.3 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 |
0.2 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.3 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.3 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.4 |
GO:0055013 |
cardiac muscle cell development(GO:0055013) |
| 0.0 |
0.6 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.1 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.0 |
0.2 |
GO:1900016 |
negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 |
0.1 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 |
0.2 |
GO:0042699 |
follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 |
0.2 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 |
0.2 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 |
0.1 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.0 |
0.4 |
GO:0006706 |
steroid catabolic process(GO:0006706) |
| 0.0 |
0.1 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 |
0.1 |
GO:0042908 |
xenobiotic transport(GO:0042908) |
| 0.0 |
0.1 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 |
0.6 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.1 |
GO:2000807 |
regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 |
0.2 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.8 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
1.0 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 |
1.3 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
0.6 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 |
0.4 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.1 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.0 |
0.1 |
GO:0046381 |
CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.0 |
0.5 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
0.1 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.0 |
0.4 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.2 |
GO:0045040 |
protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 |
0.1 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.0 |
0.2 |
GO:1904321 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 |
0.2 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 |
0.3 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.2 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 |
0.5 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.0 |
0.2 |
GO:0036120 |
cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 |
0.2 |
GO:0051770 |
positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 |
0.6 |
GO:0031103 |
axon regeneration(GO:0031103) |
| 0.0 |
0.5 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 |
0.1 |
GO:0001923 |
B-1 B cell differentiation(GO:0001923) |
| 0.0 |
0.1 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 |
0.1 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 |
0.1 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 |
0.6 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
0.2 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 |
0.5 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.6 |
GO:0045921 |
positive regulation of exocytosis(GO:0045921) |
| 0.0 |
0.2 |
GO:0072520 |
seminiferous tubule development(GO:0072520) |
| 0.0 |
0.2 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 |
0.0 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.0 |
0.3 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
0.0 |
GO:0006106 |
fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 0.0 |
0.2 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 |
0.0 |
GO:0070172 |
positive regulation of tooth mineralization(GO:0070172) |
| 0.0 |
0.0 |
GO:0034447 |
very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 |
0.4 |
GO:0090022 |
regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 |
0.1 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 |
0.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.4 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.4 |
GO:0031571 |
mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 |
0.2 |
GO:0010738 |
regulation of protein kinase A signaling(GO:0010738) |
| 0.0 |
0.1 |
GO:0009396 |
folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 |
0.0 |
GO:0031133 |
cellular magnesium ion homeostasis(GO:0010961) regulation of axon diameter(GO:0031133) |
| 0.0 |
0.1 |
GO:0030422 |
production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 |
0.4 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.1 |
GO:0097501 |
stress response to metal ion(GO:0097501) |
| 0.0 |
0.4 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.1 |
GO:0006048 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 |
0.4 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 |
0.0 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 |
0.1 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |
| 0.0 |
0.1 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 |
0.7 |
GO:0007492 |
endoderm development(GO:0007492) |
| 0.0 |
0.4 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.3 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.6 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.0 |
0.3 |
GO:0006356 |
regulation of transcription from RNA polymerase I promoter(GO:0006356) |