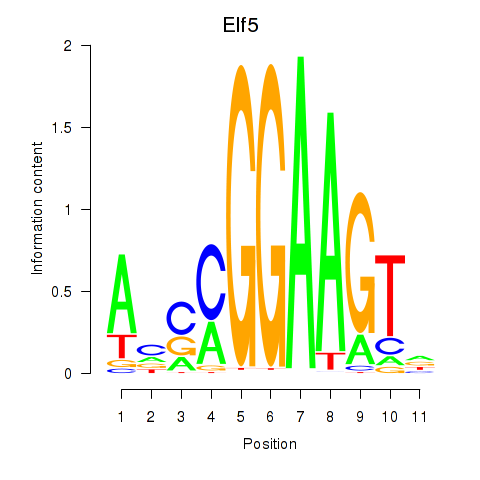
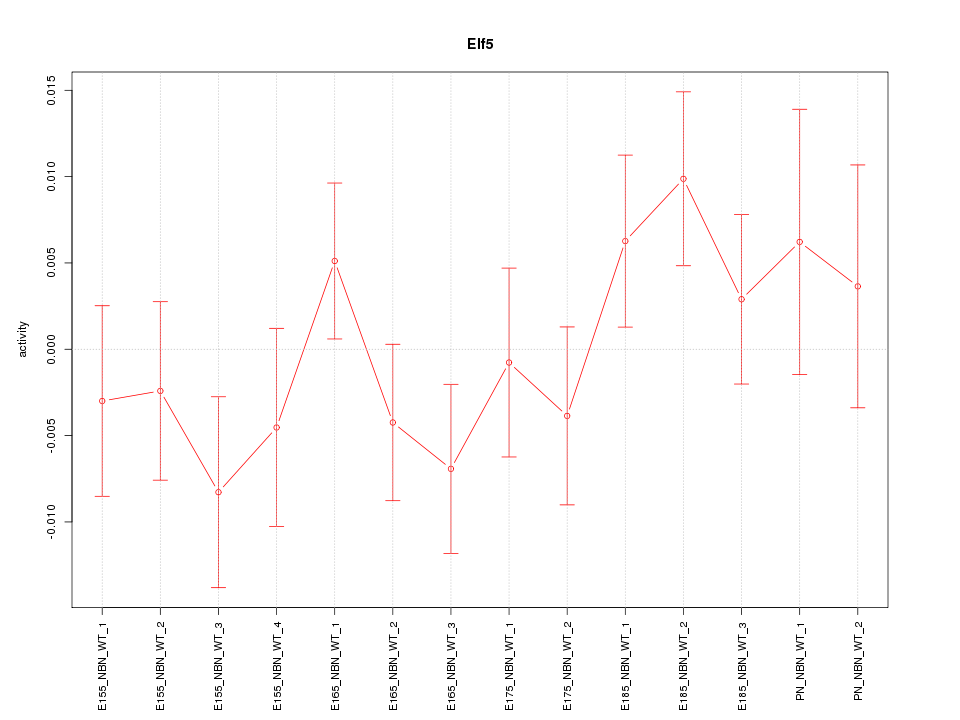
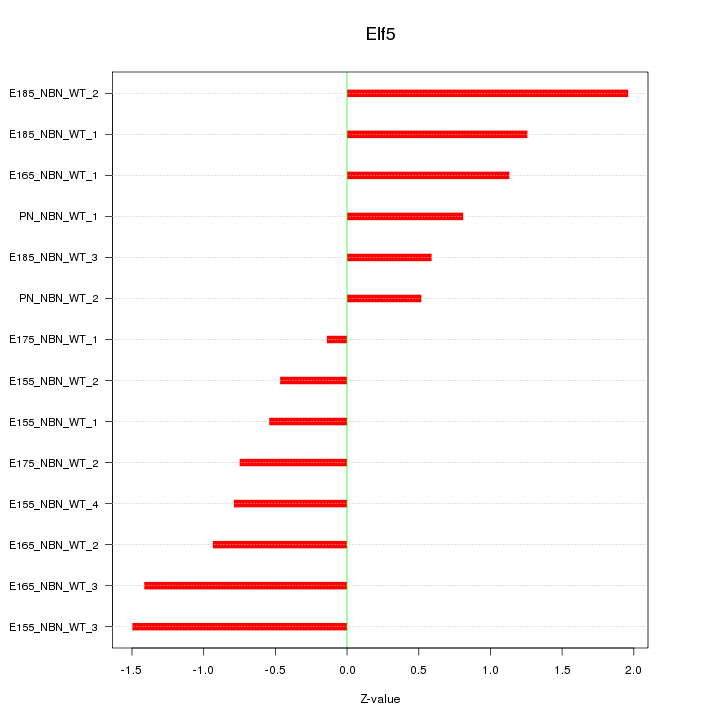
| 0.7 |
2.6 |
GO:1900426 |
positive regulation of defense response to bacterium(GO:1900426) |
| 0.6 |
1.8 |
GO:0072223 |
metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.5 |
1.5 |
GO:0090187 |
positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.5 |
1.5 |
GO:0019043 |
establishment of viral latency(GO:0019043) |
| 0.4 |
1.2 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.3 |
0.7 |
GO:0050859 |
negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.3 |
0.3 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.3 |
0.9 |
GO:0070476 |
rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.3 |
0.9 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.3 |
1.1 |
GO:0032815 |
negative regulation of natural killer cell activation(GO:0032815) |
| 0.3 |
0.3 |
GO:0043366 |
beta selection(GO:0043366) |
| 0.3 |
1.1 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 |
1.3 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.2 |
1.5 |
GO:0001302 |
replicative cell aging(GO:0001302) |
| 0.2 |
0.7 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.2 |
0.7 |
GO:0031990 |
mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.2 |
0.5 |
GO:0034135 |
regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.2 |
0.7 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) negative regulation of lymphocyte migration(GO:2000402) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.2 |
0.7 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.2 |
1.1 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.2 |
0.8 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 |
0.8 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 |
0.4 |
GO:0018171 |
peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 |
0.6 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.2 |
0.4 |
GO:2000152 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of ubiquitin-specific protease activity(GO:2000152) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.2 |
0.2 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.2 |
0.6 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.2 |
0.8 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.2 |
4.0 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.2 |
0.5 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.2 |
1.2 |
GO:0019370 |
leukotriene biosynthetic process(GO:0019370) |
| 0.2 |
0.5 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 |
0.5 |
GO:0060785 |
regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.2 |
0.7 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.2 |
0.5 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 |
0.5 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 |
0.6 |
GO:1903984 |
positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 |
0.6 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 |
0.2 |
GO:2000425 |
regulation of apoptotic cell clearance(GO:2000425) |
| 0.2 |
1.1 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 |
0.4 |
GO:0001788 |
antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.1 |
0.3 |
GO:0006296 |
nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.1 |
0.4 |
GO:1904685 |
positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.1 |
0.6 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 |
0.4 |
GO:0016256 |
N-glycan processing to lysosome(GO:0016256) |
| 0.1 |
2.1 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.1 |
0.5 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) |
| 0.1 |
1.0 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 |
0.5 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 |
1.1 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.1 |
0.1 |
GO:1902455 |
negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 |
0.5 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 |
0.3 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.1 |
0.6 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.1 |
0.4 |
GO:0003195 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 |
0.4 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 |
0.5 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 |
0.7 |
GO:0003065 |
positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 |
0.5 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 |
0.9 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.3 |
GO:0035696 |
monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.1 |
0.4 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 |
0.1 |
GO:1901798 |
positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 |
1.1 |
GO:0045651 |
positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 |
0.4 |
GO:1903181 |
regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 |
0.6 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 |
0.3 |
GO:1904569 |
regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 |
0.4 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.1 |
0.8 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.1 |
0.2 |
GO:0032701 |
negative regulation of interleukin-18 production(GO:0032701) |
| 0.1 |
0.4 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 |
0.6 |
GO:0044034 |
negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 |
0.5 |
GO:0060051 |
negative regulation of protein glycosylation(GO:0060051) |
| 0.1 |
3.6 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 |
0.4 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 |
0.3 |
GO:0098787 |
mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 |
0.2 |
GO:0010757 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) negative regulation of plasminogen activation(GO:0010757) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 |
0.6 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.1 |
0.3 |
GO:2000642 |
negative regulation of vacuolar transport(GO:1903336) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 |
0.1 |
GO:0070070 |
proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 |
0.6 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
0.3 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.1 |
0.3 |
GO:0086017 |
Purkinje myocyte action potential(GO:0086017) |
| 0.1 |
0.4 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 |
0.7 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 |
0.4 |
GO:2001270 |
regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.1 |
1.0 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
0.4 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 |
0.4 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 |
1.6 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |
| 0.1 |
0.3 |
GO:0043181 |
vacuolar sequestering(GO:0043181) |
| 0.1 |
0.2 |
GO:0002922 |
positive regulation of humoral immune response(GO:0002922) |
| 0.1 |
0.3 |
GO:0071051 |
polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 |
0.3 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.1 |
1.2 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.1 |
0.7 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.1 |
0.5 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.1 |
0.2 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.1 |
0.3 |
GO:1904749 |
regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 |
1.6 |
GO:0061298 |
retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 |
0.2 |
GO:0071038 |
U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.1 |
0.4 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 |
0.4 |
GO:0000430 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 |
1.4 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.1 |
0.5 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.1 |
0.2 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 |
0.2 |
GO:1901228 |
positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 |
0.3 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.1 |
0.3 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 |
0.3 |
GO:0009212 |
dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 |
0.5 |
GO:0030853 |
negative regulation of granulocyte differentiation(GO:0030853) |
| 0.1 |
0.2 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 |
0.2 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.3 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 |
0.3 |
GO:1902990 |
mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.1 |
0.2 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 |
0.5 |
GO:2001054 |
negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 |
0.4 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.3 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 |
0.3 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.1 |
0.3 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 |
0.1 |
GO:0010700 |
negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 |
0.5 |
GO:0090045 |
positive regulation of deacetylase activity(GO:0090045) |
| 0.1 |
0.8 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.1 |
0.1 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 0.1 |
0.1 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.1 |
0.3 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.1 |
0.3 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 |
0.2 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.1 |
0.4 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 |
0.4 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.1 |
0.3 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 |
0.2 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.1 |
0.2 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 |
0.4 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.1 |
0.1 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.1 |
0.3 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.1 |
0.4 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 |
0.8 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
0.3 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.1 |
0.1 |
GO:0010730 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 |
0.3 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.1 |
0.5 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.1 |
0.4 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.1 |
0.2 |
GO:0036451 |
cap mRNA methylation(GO:0036451) |
| 0.1 |
0.3 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 |
0.2 |
GO:0000393 |
spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 |
0.4 |
GO:1901977 |
negative regulation of cell cycle checkpoint(GO:1901977) |
| 0.1 |
0.3 |
GO:0051182 |
coenzyme transport(GO:0051182) |
| 0.1 |
0.4 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.1 |
0.2 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 |
0.2 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 |
0.1 |
GO:0021508 |
floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 |
0.2 |
GO:0045414 |
regulation of interleukin-8 biosynthetic process(GO:0045414) |
| 0.1 |
2.1 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.1 |
0.2 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.1 |
0.7 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.1 |
0.4 |
GO:0051770 |
positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.1 |
0.1 |
GO:0034472 |
snRNA 3'-end processing(GO:0034472) |
| 0.1 |
0.3 |
GO:0060576 |
intestinal epithelial cell development(GO:0060576) |
| 0.1 |
0.1 |
GO:0046070 |
dGTP metabolic process(GO:0046070) |
| 0.1 |
0.3 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 |
0.2 |
GO:0010989 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 |
0.3 |
GO:0032782 |
bile acid secretion(GO:0032782) |
| 0.1 |
0.3 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.1 |
1.4 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.1 |
0.2 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 |
0.3 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.1 |
0.9 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.1 |
0.3 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 |
0.3 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) |
| 0.1 |
0.4 |
GO:0009146 |
purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.1 |
0.4 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 |
0.3 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.1 |
0.2 |
GO:0060709 |
glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.1 |
0.3 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
0.3 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.6 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 |
0.3 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.9 |
GO:1903020 |
positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 |
0.2 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.0 |
0.6 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 |
0.2 |
GO:0034372 |
very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 |
0.7 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
0.3 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.0 |
0.2 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 |
0.1 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 |
0.3 |
GO:0035743 |
CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 |
0.2 |
GO:0032968 |
positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 |
0.1 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 |
0.2 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.0 |
0.5 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 |
0.8 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.0 |
0.2 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 |
0.3 |
GO:0070863 |
positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 |
0.2 |
GO:0010755 |
regulation of plasminogen activation(GO:0010755) |
| 0.0 |
0.9 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
0.7 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.3 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.1 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 |
1.4 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.1 |
GO:0071335 |
hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 |
0.9 |
GO:0050829 |
defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 |
0.2 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.0 |
0.2 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 |
0.1 |
GO:0014891 |
striated muscle atrophy(GO:0014891) |
| 0.0 |
0.6 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 |
0.3 |
GO:0010561 |
negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 |
0.9 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.2 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
0.4 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.1 |
GO:0035880 |
embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 |
0.5 |
GO:0032515 |
negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 |
0.1 |
GO:2000015 |
regulation of determination of dorsal identity(GO:2000015) |
| 0.0 |
0.3 |
GO:0070474 |
positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.0 |
0.3 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 |
0.0 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.0 |
0.0 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 |
2.2 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 |
0.2 |
GO:0090219 |
negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 |
0.2 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.6 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) |
| 0.0 |
0.2 |
GO:0035624 |
mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) response to high density lipoprotein particle(GO:0055099) |
| 0.0 |
0.6 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.8 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
1.5 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.0 |
0.0 |
GO:0002679 |
respiratory burst involved in defense response(GO:0002679) |
| 0.0 |
0.1 |
GO:0060753 |
regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 |
0.2 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of nuclease activity(GO:0032074) negative regulation of immunoglobulin secretion(GO:0051025) regulation of endoribonuclease activity(GO:0060699) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 |
0.2 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.0 |
0.6 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 |
0.1 |
GO:0042726 |
flavin-containing compound metabolic process(GO:0042726) |
| 0.0 |
0.6 |
GO:2001241 |
positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 |
0.2 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 |
0.8 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.2 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 |
0.3 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.4 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.2 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 |
0.4 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.3 |
GO:2000645 |
negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 |
0.2 |
GO:0006046 |
N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 |
0.1 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.0 |
0.3 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.2 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.0 |
0.2 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.2 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 |
0.2 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.4 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.3 |
GO:0032096 |
negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 |
0.1 |
GO:0010248 |
B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 |
0.5 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 |
0.1 |
GO:1901301 |
regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 |
0.0 |
GO:0051643 |
endoplasmic reticulum localization(GO:0051643) |
| 0.0 |
0.1 |
GO:0051798 |
positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 |
0.3 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 |
0.2 |
GO:2000650 |
negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 |
0.2 |
GO:0021905 |
pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) positive regulation of cell fate specification(GO:0042660) positive regulation of glucagon secretion(GO:0070094) |
| 0.0 |
0.1 |
GO:0051582 |
positive regulation of neurotransmitter uptake(GO:0051582) regulation of dopamine uptake involved in synaptic transmission(GO:0051584) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 |
0.4 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.0 |
0.1 |
GO:0048319 |
axial mesoderm morphogenesis(GO:0048319) |
| 0.0 |
0.7 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.9 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.2 |
GO:0002484 |
antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.0 |
0.0 |
GO:0002309 |
T cell proliferation involved in immune response(GO:0002309) |
| 0.0 |
1.0 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.1 |
GO:0031052 |
programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.0 |
0.2 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.1 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 |
0.2 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 |
0.3 |
GO:0032688 |
negative regulation of interferon-beta production(GO:0032688) |
| 0.0 |
0.3 |
GO:0070200 |
establishment of protein localization to telomere(GO:0070200) |
| 0.0 |
0.3 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.1 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 |
0.3 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.0 |
0.1 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) |
| 0.0 |
0.1 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 |
0.4 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.1 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
0.4 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.2 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 |
0.5 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.2 |
GO:0060628 |
regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 |
0.1 |
GO:1902255 |
positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 |
0.6 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.1 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.0 |
0.0 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.0 |
0.3 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.0 |
0.1 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.0 |
0.1 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.1 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.0 |
0.1 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 |
0.3 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.0 |
0.1 |
GO:0090201 |
negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 |
0.1 |
GO:0033030 |
negative regulation of neutrophil apoptotic process(GO:0033030) metanephric tubule morphogenesis(GO:0072173) |
| 0.0 |
0.2 |
GO:0002192 |
IRES-dependent translational initiation(GO:0002192) |
| 0.0 |
0.6 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.2 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 |
0.1 |
GO:0033499 |
galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.0 |
0.2 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.1 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 |
0.6 |
GO:0006301 |
postreplication repair(GO:0006301) |
| 0.0 |
1.0 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.2 |
GO:0007623 |
circadian rhythm(GO:0007623) |
| 0.0 |
0.2 |
GO:0021554 |
optic nerve development(GO:0021554) |
| 0.0 |
0.2 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.0 |
0.1 |
GO:0009223 |
pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 |
0.1 |
GO:0048007 |
antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 |
0.4 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.0 |
GO:0001844 |
protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.0 |
0.1 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.0 |
0.1 |
GO:0061101 |
neuroendocrine cell differentiation(GO:0061101) |
| 0.0 |
0.7 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
0.1 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 |
0.2 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 |
0.4 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.2 |
GO:0071493 |
cellular response to antibiotic(GO:0071236) cellular response to UV-B(GO:0071493) |
| 0.0 |
0.2 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.0 |
0.2 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.0 |
0.1 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.2 |
GO:0006221 |
pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 |
0.1 |
GO:0070475 |
rRNA base methylation(GO:0070475) |
| 0.0 |
0.1 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.1 |
GO:0006273 |
lagging strand elongation(GO:0006273) |
| 0.0 |
0.2 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
0.2 |
GO:2000505 |
regulation of energy homeostasis(GO:2000505) |
| 0.0 |
0.2 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.0 |
0.2 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.3 |
GO:0009081 |
branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.2 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
0.1 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 |
0.2 |
GO:0035999 |
tetrahydrofolate interconversion(GO:0035999) |
| 0.0 |
0.1 |
GO:0016139 |
glycoside catabolic process(GO:0016139) |
| 0.0 |
0.4 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.0 |
0.1 |
GO:0042136 |
neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 |
0.2 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.4 |
GO:0055012 |
ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 |
0.6 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.2 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.3 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
0.0 |
GO:0061724 |
lipophagy(GO:0061724) |
| 0.0 |
0.0 |
GO:0070827 |
chromatin maintenance(GO:0070827) heterochromatin maintenance(GO:0070829) |
| 0.0 |
0.1 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.0 |
0.1 |
GO:0035457 |
cellular response to interferon-alpha(GO:0035457) |
| 0.0 |
0.1 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.2 |
GO:0043277 |
apoptotic cell clearance(GO:0043277) |
| 0.0 |
0.0 |
GO:0098501 |
DNA damage response, detection of DNA damage(GO:0042769) polynucleotide dephosphorylation(GO:0098501) |
| 0.0 |
0.2 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.2 |
GO:0051898 |
negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 |
0.2 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 |
0.0 |
GO:0043388 |
positive regulation of DNA binding(GO:0043388) |
| 0.0 |
0.6 |
GO:0009142 |
nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 |
0.4 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.1 |
GO:1901164 |
negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 |
0.1 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.1 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.0 |
0.1 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.2 |
GO:0010867 |
positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 |
0.1 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 |
0.2 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.3 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.1 |
GO:0009651 |
response to salt stress(GO:0009651) |
| 0.0 |
0.3 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.2 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.1 |
GO:0002634 |
regulation of germinal center formation(GO:0002634) |
| 0.0 |
0.1 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.0 |
0.1 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.0 |
GO:0042940 |
D-amino acid transport(GO:0042940) |
| 0.0 |
0.4 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.1 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.0 |
GO:0002361 |
CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 |
0.1 |
GO:0043173 |
purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) nucleotide salvage(GO:0043173) |
| 0.0 |
0.1 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.1 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 |
0.1 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.1 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 |
0.0 |
GO:0034184 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.2 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 |
0.1 |
GO:0039703 |
viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 |
0.0 |
GO:0031145 |
anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 |
0.0 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 |
0.1 |
GO:1903963 |
arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 |
0.0 |
GO:0010626 |
negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 |
0.1 |
GO:0044144 |
regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 |
0.1 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.1 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.2 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.0 |
GO:0042713 |
sperm ejaculation(GO:0042713) |
| 0.0 |
0.0 |
GO:0044849 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) estrous cycle(GO:0044849) |
| 0.0 |
0.0 |
GO:0042045 |
epithelial fluid transport(GO:0042045) |
| 0.0 |
0.1 |
GO:0051123 |
RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 |
0.1 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.0 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 |
0.2 |
GO:0051351 |
positive regulation of ligase activity(GO:0051351) |
| 0.0 |
0.1 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
0.3 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.0 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 |
0.1 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.2 |
GO:0043171 |
peptide catabolic process(GO:0043171) |