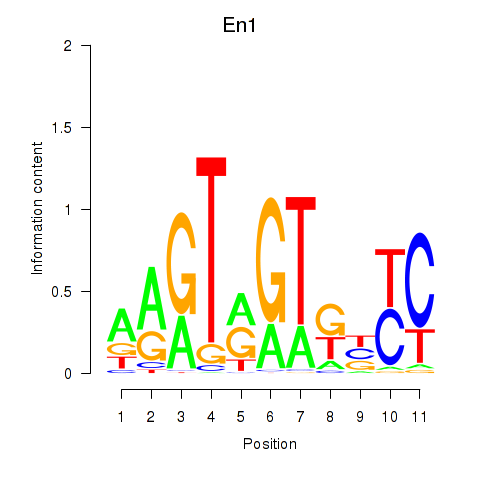
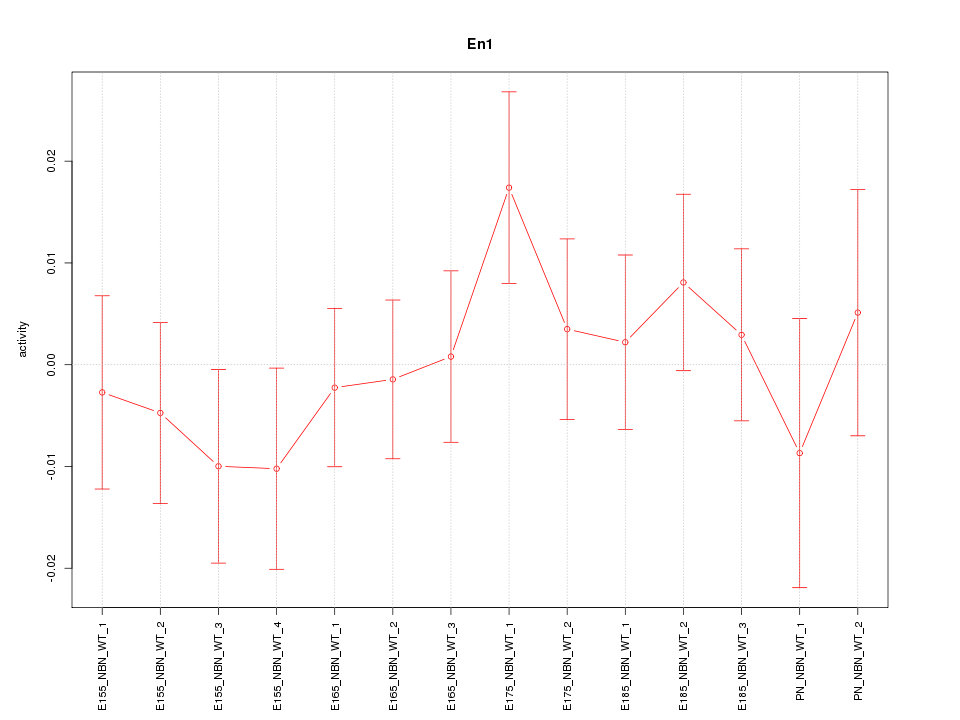
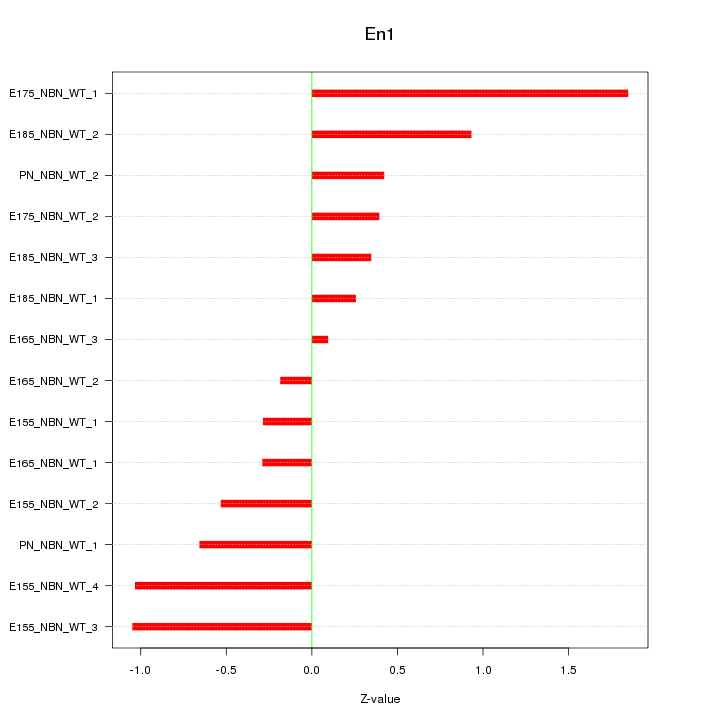
| 0.4 |
2.1 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.3 |
1.4 |
GO:2000065 |
negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.3 |
0.8 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.3 |
0.8 |
GO:2000170 |
positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 |
1.8 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.2 |
1.3 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.2 |
0.6 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
0.4 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 |
0.8 |
GO:0010694 |
positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.1 |
0.4 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 |
0.4 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 |
0.7 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 |
0.5 |
GO:0070295 |
renal water absorption(GO:0070295) |
| 0.1 |
0.8 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.1 |
0.3 |
GO:2000987 |
positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 |
0.3 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 |
0.3 |
GO:0032788 |
saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.1 |
0.2 |
GO:0003195 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 |
0.2 |
GO:0045410 |
positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.1 |
0.5 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.1 |
0.3 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.1 |
0.3 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.1 |
1.9 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.1 |
0.4 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
0.2 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.1 |
0.7 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.2 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 |
0.2 |
GO:0072425 |
signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 |
0.4 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 |
0.3 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.1 |
0.2 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.1 |
0.2 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.1 |
0.2 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 |
1.6 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.1 |
0.2 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 |
0.2 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 |
1.3 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.4 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.9 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
0.2 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.6 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
0.4 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.0 |
0.1 |
GO:1900623 |
positive regulation of keratinocyte proliferation(GO:0010838) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 |
0.4 |
GO:0044539 |
long-chain fatty acid import(GO:0044539) |
| 0.0 |
0.3 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.2 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 |
0.8 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.1 |
GO:0051852 |
antifungal humoral response(GO:0019732) disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 |
0.2 |
GO:0046645 |
positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 |
0.1 |
GO:0097105 |
presynaptic membrane assembly(GO:0097105) |
| 0.0 |
0.1 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 |
0.1 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 |
0.1 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 |
0.1 |
GO:0030862 |
neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.0 |
0.2 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) |
| 0.0 |
0.3 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.6 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.0 |
0.8 |
GO:0003016 |
respiratory system process(GO:0003016) |
| 0.0 |
0.1 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.0 |
0.2 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.4 |
GO:0032291 |
central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 |
0.1 |
GO:0002538 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) olefin metabolic process(GO:1900673) |
| 0.0 |
0.2 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.1 |
GO:0010636 |
positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 |
0.1 |
GO:0042097 |
interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 |
0.5 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.0 |
0.1 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 |
0.1 |
GO:0070814 |
hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 |
0.1 |
GO:0060423 |
limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) negative regulation of mitotic cell cycle, embryonic(GO:0045976) foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.0 |
0.2 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.1 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 |
0.7 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:0003418 |
growth plate cartilage chondrocyte differentiation(GO:0003418) growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 |
0.1 |
GO:0039692 |
single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) |
| 0.0 |
0.1 |
GO:0002314 |
germinal center B cell differentiation(GO:0002314) |
| 0.0 |
0.1 |
GO:0048023 |
positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 |
0.3 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.6 |
GO:0048873 |
homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 |
0.1 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.1 |
GO:1902177 |
positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 |
0.3 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.0 |
GO:0009812 |
flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 |
0.2 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 |
0.0 |
GO:0031622 |
positive regulation of fever generation(GO:0031622) |
| 0.0 |
0.1 |
GO:0010920 |
negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.0 |
0.1 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 |
0.1 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 |
0.3 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 |
0.2 |
GO:0071357 |
negative regulation of viral transcription(GO:0032897) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 |
0.5 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.1 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.3 |
GO:0090129 |
positive regulation of synapse maturation(GO:0090129) |
| 0.0 |
0.1 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.0 |
0.0 |
GO:0089700 |
protein kinase D signaling(GO:0089700) |
| 0.0 |
0.1 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 |
0.1 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.1 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.0 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |