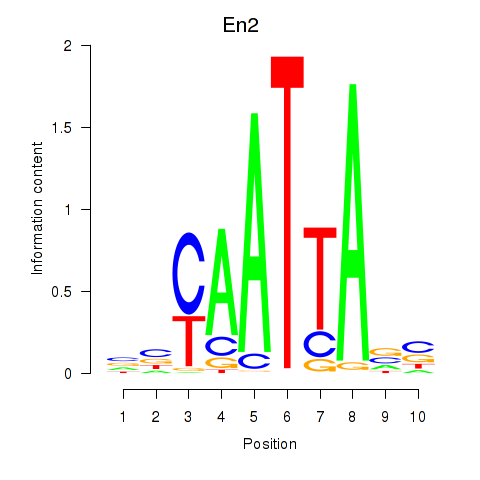
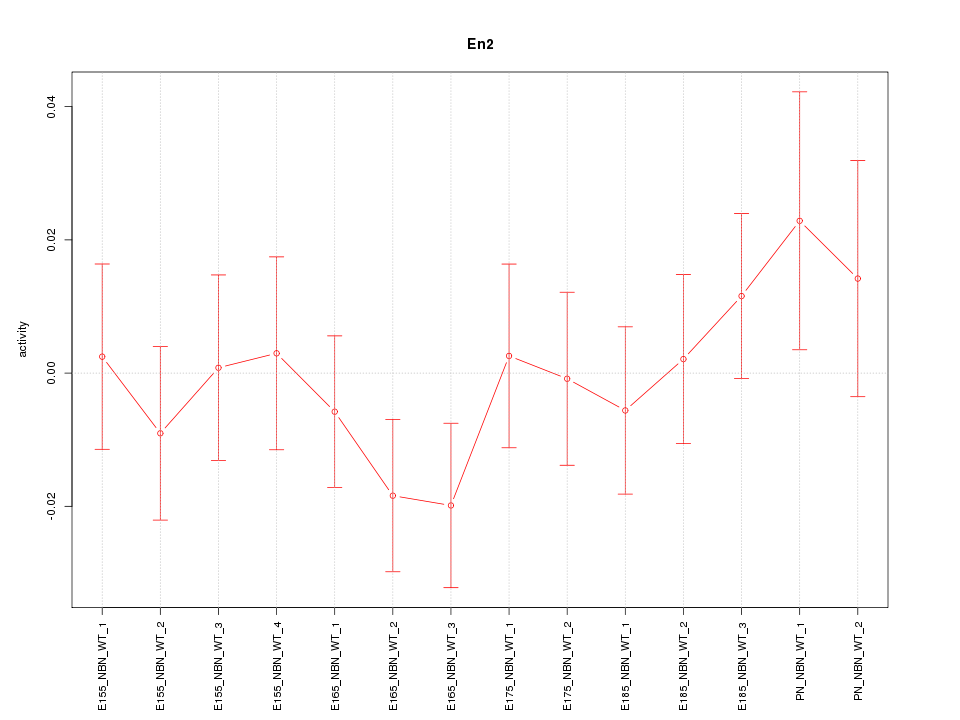
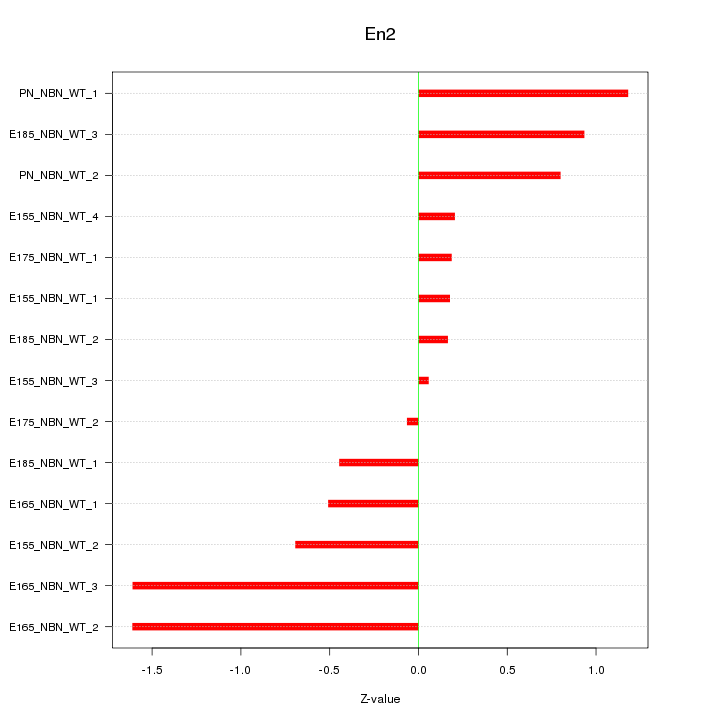
| 0.4 |
1.6 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.4 |
1.2 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.4 |
1.2 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.3 |
1.7 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.2 |
0.7 |
GO:0051342 |
regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.2 |
3.0 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 |
1.0 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 |
0.5 |
GO:0072204 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.2 |
0.9 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.1 |
0.8 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
0.4 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) regulation of vascular wound healing(GO:0061043) glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.1 |
0.5 |
GO:0072592 |
regulation of integrin biosynthetic process(GO:0045113) oxygen metabolic process(GO:0072592) |
| 0.1 |
0.3 |
GO:2000564 |
CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.1 |
1.8 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 |
0.3 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.1 |
0.3 |
GO:0090324 |
negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 |
1.4 |
GO:0030953 |
astral microtubule organization(GO:0030953) |
| 0.1 |
0.6 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.1 |
0.3 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 |
0.3 |
GO:0071475 |
cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 |
1.3 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.1 |
0.2 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 |
0.6 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.1 |
0.9 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 |
0.2 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 |
1.7 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.1 |
1.7 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 |
0.3 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 |
0.8 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.9 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.5 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
0.3 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
0.5 |
GO:0042761 |
very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 |
0.4 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 |
0.8 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
0.9 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.2 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.0 |
0.1 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 |
1.0 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.1 |
GO:1902915 |
negative regulation of protein import into nucleus, translocation(GO:0033159) negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 |
2.2 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.2 |
GO:0002361 |
CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 |
0.8 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.4 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.9 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.3 |
GO:0001973 |
adenosine receptor signaling pathway(GO:0001973) |
| 0.0 |
1.1 |
GO:0035307 |
positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 |
0.4 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.1 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 |
0.6 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.3 |
GO:0046548 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) retinal rod cell development(GO:0046548) |
| 0.0 |
0.7 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.6 |
GO:0071385 |
cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 |
0.2 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.1 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 |
1.8 |
GO:0001889 |
liver development(GO:0001889) |
| 0.0 |
0.1 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.3 |
GO:0036342 |
post-anal tail morphogenesis(GO:0036342) |
| 0.0 |
0.3 |
GO:0061157 |
mRNA destabilization(GO:0061157) |
| 0.0 |
0.2 |
GO:0099625 |
regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.0 |
0.5 |
GO:0006801 |
superoxide metabolic process(GO:0006801) |
| 0.0 |
0.2 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |