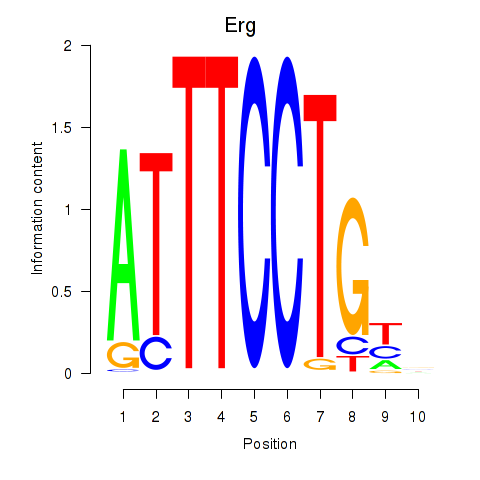
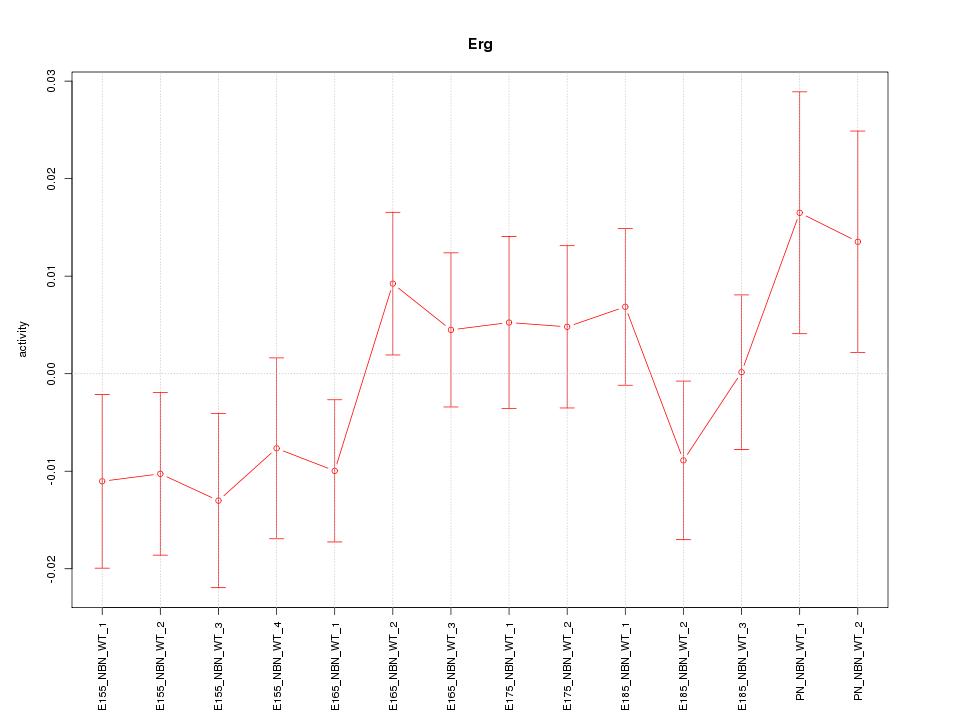
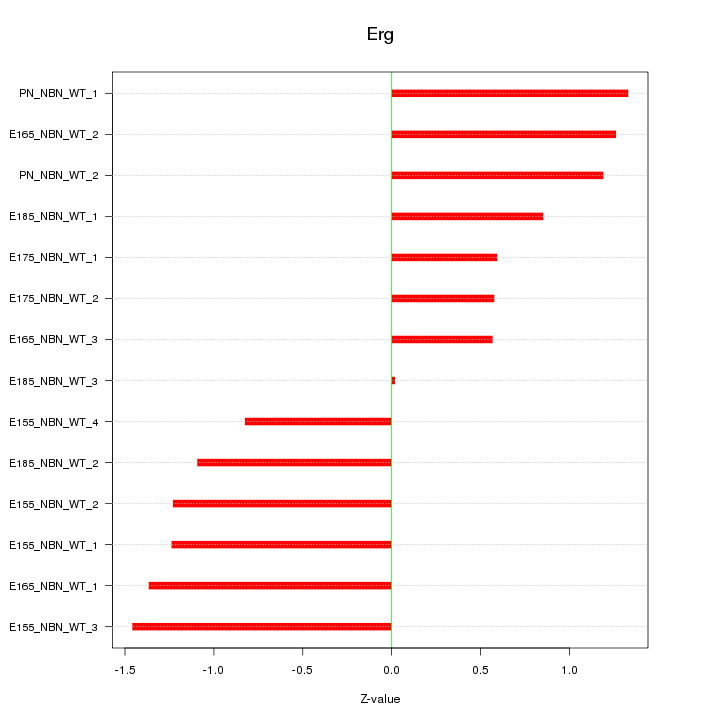
| 0.7 |
2.2 |
GO:0003032 |
detection of oxygen(GO:0003032) |
| 0.6 |
7.0 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.6 |
3.1 |
GO:2001204 |
regulation of osteoclast development(GO:2001204) |
| 0.6 |
1.8 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.6 |
1.7 |
GO:0090187 |
zymogen granule exocytosis(GO:0070625) positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.4 |
2.1 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.4 |
1.1 |
GO:0050859 |
negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.4 |
1.4 |
GO:1900272 |
negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.3 |
2.0 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.3 |
1.0 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.3 |
1.0 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.3 |
1.2 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.3 |
0.8 |
GO:0021649 |
vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.2 |
0.7 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.2 |
2.1 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 |
2.3 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 |
0.9 |
GO:0061010 |
gall bladder development(GO:0061010) |
| 0.2 |
0.9 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.2 |
0.9 |
GO:0006447 |
regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 |
0.9 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.2 |
1.1 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 |
2.0 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 |
0.4 |
GO:1901492 |
positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 |
1.4 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.2 |
1.0 |
GO:0060842 |
arterial endothelial cell differentiation(GO:0060842) |
| 0.2 |
1.5 |
GO:1901534 |
positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.2 |
1.0 |
GO:0071638 |
negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 |
1.5 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.2 |
0.8 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 |
2.7 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.2 |
1.4 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.2 |
0.8 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.2 |
2.0 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 |
2.7 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.1 |
0.4 |
GO:0001803 |
antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.1 |
0.5 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
1.0 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.1 |
0.5 |
GO:1904192 |
prostate gland stromal morphogenesis(GO:0060741) cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 |
0.9 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.6 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 |
0.8 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
0.1 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 |
0.3 |
GO:0035880 |
embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 |
0.4 |
GO:2000313 |
fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 |
0.1 |
GO:0042119 |
neutrophil activation involved in immune response(GO:0002283) neutrophil activation(GO:0042119) |
| 0.1 |
0.6 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.1 |
0.4 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 |
0.4 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.1 |
0.6 |
GO:0002681 |
somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 |
0.3 |
GO:0035435 |
phosphate ion transmembrane transport(GO:0035435) |
| 0.1 |
0.7 |
GO:0016560 |
protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 |
0.5 |
GO:1901164 |
negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 |
0.3 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 |
0.4 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.3 |
GO:0031086 |
nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 |
0.3 |
GO:2001180 |
regulation of interleukin-18 production(GO:0032661) negative regulation of interleukin-18 production(GO:0032701) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.1 |
0.8 |
GO:0042226 |
interleukin-6 biosynthetic process(GO:0042226) regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.1 |
0.2 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 |
0.3 |
GO:0043652 |
engulfment of apoptotic cell(GO:0043652) |
| 0.1 |
0.4 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.1 |
0.7 |
GO:2001053 |
regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 |
0.1 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.1 |
0.2 |
GO:0002322 |
B cell proliferation involved in immune response(GO:0002322) |
| 0.1 |
2.5 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.1 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 |
0.3 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 |
0.9 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.1 |
0.1 |
GO:1900020 |
regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 |
0.4 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.3 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 |
0.4 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.1 |
0.1 |
GO:0060486 |
Clara cell differentiation(GO:0060486) |
| 0.1 |
0.2 |
GO:0042637 |
catagen(GO:0042637) |
| 0.1 |
0.4 |
GO:2000002 |
negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 |
0.4 |
GO:0045651 |
positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 |
0.2 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 |
0.6 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 |
0.3 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.1 |
0.2 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 |
0.4 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 |
0.2 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 |
0.7 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 |
1.0 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.0 |
3.8 |
GO:0016525 |
negative regulation of angiogenesis(GO:0016525) |
| 0.0 |
3.2 |
GO:2001238 |
positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 |
0.7 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.3 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
0.9 |
GO:0030947 |
regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 |
0.6 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 |
1.0 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.0 |
0.3 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.2 |
GO:0014835 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 |
0.4 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.6 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.0 |
0.2 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 |
0.4 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.0 |
0.1 |
GO:2000619 |
negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 |
0.8 |
GO:0034340 |
response to type I interferon(GO:0034340) |
| 0.0 |
1.4 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
0.3 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.3 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.8 |
GO:0035458 |
cellular response to interferon-beta(GO:0035458) |
| 0.0 |
0.3 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.0 |
0.1 |
GO:0098501 |
polynucleotide dephosphorylation(GO:0098501) |
| 0.0 |
0.2 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 |
1.4 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.1 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
1.1 |
GO:0051567 |
histone H3-K9 methylation(GO:0051567) |
| 0.0 |
0.3 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.0 |
0.3 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
1.4 |
GO:0017145 |
stem cell division(GO:0017145) |
| 0.0 |
1.1 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.3 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.0 |
0.1 |
GO:0036438 |
positive regulation of heterotypic cell-cell adhesion(GO:0034116) maintenance of lens transparency(GO:0036438) |
| 0.0 |
0.1 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 |
0.3 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.0 |
GO:0010936 |
negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 |
0.1 |
GO:0019043 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) establishment of viral latency(GO:0019043) |
| 0.0 |
0.6 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.1 |
GO:0060449 |
negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) bud elongation involved in lung branching(GO:0060449) |
| 0.0 |
0.1 |
GO:0042364 |
water-soluble vitamin biosynthetic process(GO:0042364) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 |
0.1 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.0 |
0.3 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.5 |
GO:0035066 |
positive regulation of histone acetylation(GO:0035066) |
| 0.0 |
0.4 |
GO:0043486 |
histone exchange(GO:0043486) |
| 0.0 |
0.0 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 |
0.8 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.0 |
0.3 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.0 |
0.1 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.0 |
0.1 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 |
0.1 |
GO:0006625 |
protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 |
0.2 |
GO:0023041 |
neuronal signal transduction(GO:0023041) |
| 0.0 |
0.1 |
GO:0051231 |
mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 |
0.1 |
GO:1901857 |
positive regulation of cellular respiration(GO:1901857) |
| 0.0 |
0.5 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.3 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 |
0.2 |
GO:0030502 |
negative regulation of bone mineralization(GO:0030502) |
| 0.0 |
0.1 |
GO:1903215 |
regulation of mRNA modification(GO:0090365) negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 |
0.4 |
GO:0060004 |
reflex(GO:0060004) |
| 0.0 |
1.0 |
GO:0016574 |
histone ubiquitination(GO:0016574) |
| 0.0 |
0.8 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
0.7 |
GO:0051496 |
positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 |
0.3 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.1 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
0.3 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.0 |
0.2 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |
| 0.0 |
0.1 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 |
0.7 |
GO:2000649 |
regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 |
0.1 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 |
0.5 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.0 |
0.2 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.1 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.1 |
GO:0090085 |
regulation of protein deubiquitination(GO:0090085) |
| 0.0 |
0.1 |
GO:0044854 |
plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 |
0.5 |
GO:0051591 |
response to cAMP(GO:0051591) |
| 0.0 |
0.1 |
GO:0006560 |
proline metabolic process(GO:0006560) |
| 0.0 |
1.2 |
GO:0014013 |
regulation of gliogenesis(GO:0014013) |
| 0.0 |
0.0 |
GO:1901069 |
guanosine-containing compound catabolic process(GO:1901069) |