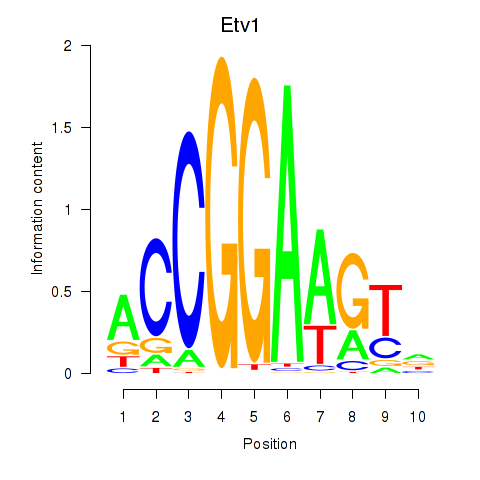
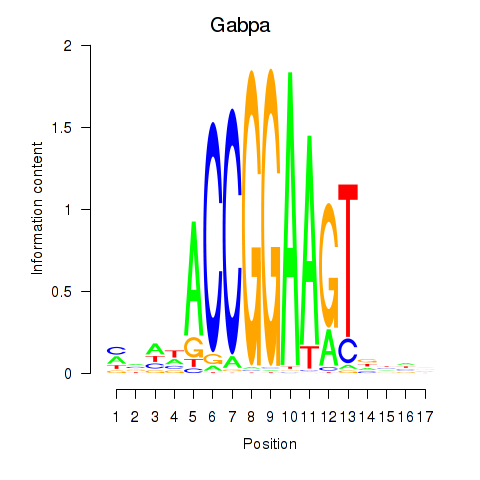
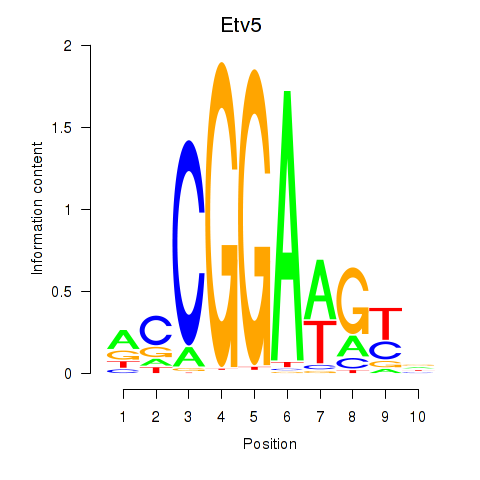
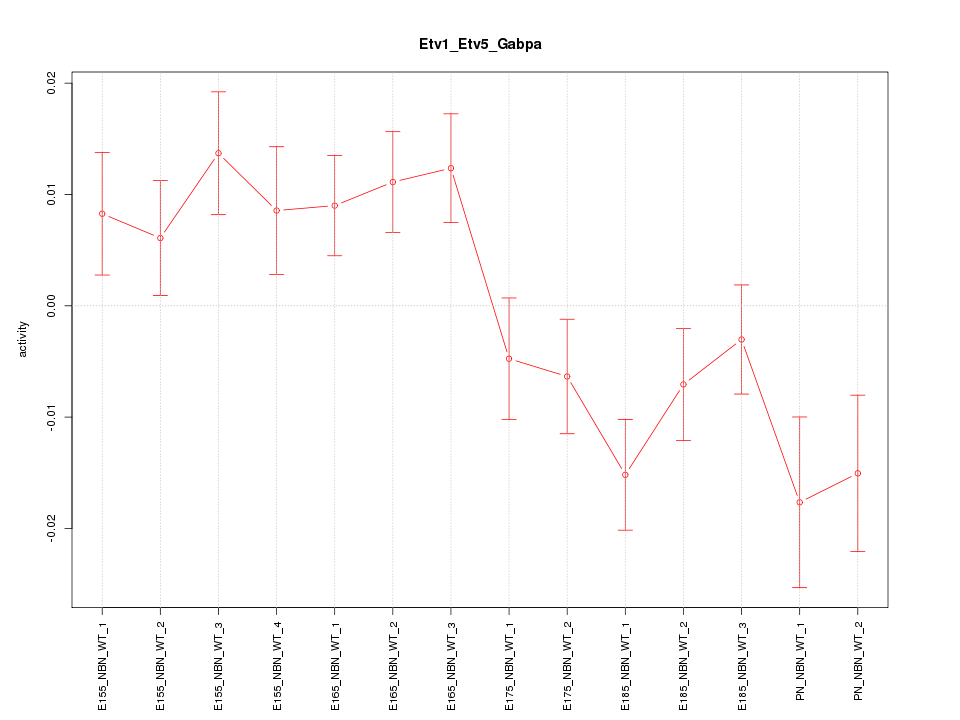
| 1.6 |
4.7 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 1.5 |
4.6 |
GO:1903896 |
positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 1.4 |
4.1 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 1.2 |
5.0 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.9 |
2.7 |
GO:0090071 |
negative regulation of ribosome biogenesis(GO:0090071) |
| 0.9 |
0.9 |
GO:0060903 |
regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of reciprocal meiotic recombination(GO:0010845) regulation of meiosis I(GO:0060631) positive regulation of meiosis I(GO:0060903) |
| 0.8 |
2.5 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.8 |
2.4 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.8 |
2.3 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.8 |
2.3 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.7 |
2.9 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.7 |
2.1 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.7 |
3.5 |
GO:0002024 |
diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.7 |
2.0 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.7 |
3.4 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.6 |
1.8 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.6 |
2.4 |
GO:0014042 |
positive regulation of neuron maturation(GO:0014042) |
| 0.6 |
1.7 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.6 |
2.3 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.6 |
1.7 |
GO:0070124 |
mitochondrial translational initiation(GO:0070124) |
| 0.6 |
1.7 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.6 |
2.3 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.6 |
0.6 |
GO:0036260 |
7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.5 |
2.1 |
GO:0010796 |
regulation of multivesicular body size(GO:0010796) |
| 0.5 |
2.5 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.5 |
1.5 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.5 |
2.4 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.5 |
1.9 |
GO:0061511 |
centriole elongation(GO:0061511) |
| 0.5 |
1.4 |
GO:0031086 |
nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.5 |
3.8 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.5 |
1.4 |
GO:0016344 |
meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.5 |
4.6 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.5 |
2.7 |
GO:0006370 |
7-methylguanosine mRNA capping(GO:0006370) |
| 0.4 |
3.5 |
GO:0000467 |
exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.4 |
1.7 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.4 |
4.3 |
GO:1900364 |
negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.4 |
1.3 |
GO:0048611 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.4 |
1.2 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.4 |
1.2 |
GO:0036228 |
protein targeting to nuclear inner membrane(GO:0036228) |
| 0.4 |
1.2 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.4 |
1.2 |
GO:1902605 |
heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.4 |
1.2 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.4 |
2.7 |
GO:0006701 |
progesterone biosynthetic process(GO:0006701) |
| 0.4 |
1.1 |
GO:1905065 |
hematopoietic stem cell migration(GO:0035701) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.4 |
4.5 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.4 |
1.9 |
GO:1900017 |
positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.4 |
1.9 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.4 |
2.2 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.4 |
1.1 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.4 |
2.9 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.4 |
1.8 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 |
2.0 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.3 |
0.3 |
GO:0016078 |
tRNA catabolic process(GO:0016078) |
| 0.3 |
0.3 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.3 |
5.0 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.3 |
2.0 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.3 |
1.0 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.3 |
1.3 |
GO:0038108 |
negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.3 |
1.0 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.3 |
2.3 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.3 |
1.3 |
GO:0070086 |
ubiquitin-dependent endocytosis(GO:0070086) |
| 0.3 |
5.1 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.3 |
1.3 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.3 |
1.2 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.3 |
0.6 |
GO:0090160 |
Golgi to lysosome transport(GO:0090160) |
| 0.3 |
0.9 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.3 |
1.5 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.3 |
3.7 |
GO:0035646 |
endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.3 |
0.9 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.3 |
0.9 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.3 |
0.9 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 |
3.2 |
GO:0097151 |
positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.3 |
2.3 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 |
0.9 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.3 |
0.6 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.3 |
3.5 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.3 |
1.1 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.3 |
1.1 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.3 |
0.8 |
GO:0051030 |
snRNA transport(GO:0051030) |
| 0.3 |
0.8 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.3 |
0.8 |
GO:0000710 |
meiotic mismatch repair(GO:0000710) |
| 0.3 |
0.8 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 |
2.1 |
GO:2000035 |
regulation of stem cell division(GO:2000035) |
| 0.3 |
1.0 |
GO:0061626 |
pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.3 |
1.3 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.3 |
1.3 |
GO:0018202 |
peptidyl-histidine modification(GO:0018202) |
| 0.2 |
0.7 |
GO:1990046 |
positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) positive regulation of mitochondrial DNA metabolic process(GO:1901860) stress-induced mitochondrial fusion(GO:1990046) |
| 0.2 |
3.7 |
GO:0009191 |
ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.2 |
2.5 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 |
4.4 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.2 |
1.2 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 |
1.0 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.2 |
1.0 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 |
2.6 |
GO:0080009 |
mRNA methylation(GO:0080009) |
| 0.2 |
1.2 |
GO:0090234 |
regulation of kinetochore assembly(GO:0090234) |
| 0.2 |
4.5 |
GO:1902236 |
negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.2 |
5.1 |
GO:0030033 |
microvillus assembly(GO:0030033) |
| 0.2 |
0.7 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.2 |
2.5 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.2 |
0.7 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 |
0.9 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.2 |
4.2 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.2 |
0.7 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.2 |
0.7 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 |
0.6 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 |
1.3 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 |
2.1 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.2 |
0.4 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.2 |
0.8 |
GO:0086042 |
cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.2 |
1.4 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.2 |
0.6 |
GO:0071500 |
cellular response to nitrosative stress(GO:0071500) |
| 0.2 |
0.8 |
GO:2000252 |
negative regulation of feeding behavior(GO:2000252) |
| 0.2 |
0.4 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 |
2.2 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 |
0.4 |
GO:0032055 |
negative regulation of translation in response to stress(GO:0032055) |
| 0.2 |
1.0 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 |
1.0 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.2 |
0.6 |
GO:0008594 |
photoreceptor cell morphogenesis(GO:0008594) |
| 0.2 |
1.4 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.2 |
1.0 |
GO:0061084 |
negative regulation of protein refolding(GO:0061084) |
| 0.2 |
1.2 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.2 |
1.0 |
GO:0060398 |
regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.2 |
0.2 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.2 |
0.6 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 |
0.2 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.2 |
0.8 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.2 |
0.2 |
GO:0002925 |
positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.2 |
1.1 |
GO:0030223 |
neutrophil differentiation(GO:0030223) |
| 0.2 |
0.9 |
GO:0060124 |
positive regulation of growth hormone secretion(GO:0060124) |
| 0.2 |
0.4 |
GO:1903599 |
positive regulation of mitophagy(GO:1903599) |
| 0.2 |
1.3 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.2 |
1.1 |
GO:0001927 |
exocyst assembly(GO:0001927) |
| 0.2 |
0.2 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.2 |
0.7 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 |
0.5 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.2 |
1.1 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 |
4.2 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 |
0.7 |
GO:0002309 |
T cell proliferation involved in immune response(GO:0002309) |
| 0.2 |
0.5 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.2 |
0.7 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.2 |
0.5 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 |
0.9 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.2 |
0.9 |
GO:0015871 |
choline transport(GO:0015871) synaptic vesicle targeting(GO:0016080) regulation of resting membrane potential(GO:0060075) |
| 0.2 |
0.5 |
GO:0000965 |
mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.2 |
0.2 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.2 |
0.5 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.2 |
0.3 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.2 |
3.8 |
GO:0051654 |
establishment of mitochondrion localization(GO:0051654) |
| 0.2 |
1.3 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.2 |
0.3 |
GO:0090611 |
ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.2 |
0.7 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.2 |
1.8 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.2 |
0.8 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.2 |
0.5 |
GO:0000454 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 |
1.1 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 |
1.8 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.2 |
1.8 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.2 |
0.3 |
GO:0035928 |
rRNA import into mitochondrion(GO:0035928) |
| 0.2 |
2.1 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 |
0.6 |
GO:0071105 |
response to interleukin-11(GO:0071105) osteoclast fusion(GO:0072675) |
| 0.2 |
1.3 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 |
0.5 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 |
0.6 |
GO:1901096 |
regulation of autophagosome maturation(GO:1901096) |
| 0.2 |
0.2 |
GO:0036480 |
neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 |
1.6 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.2 |
0.5 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.2 |
0.8 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.1 |
0.4 |
GO:0030647 |
polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.1 |
0.6 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.1 |
2.2 |
GO:0071801 |
regulation of podosome assembly(GO:0071801) |
| 0.1 |
0.6 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.1 |
0.7 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 |
0.7 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.1 |
2.5 |
GO:0060416 |
response to growth hormone(GO:0060416) |
| 0.1 |
0.7 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.1 |
1.3 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
1.3 |
GO:0070897 |
DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.1 |
1.0 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 |
0.9 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 |
2.0 |
GO:0060013 |
righting reflex(GO:0060013) |
| 0.1 |
0.6 |
GO:0065001 |
negative regulation of histone H3-K36 methylation(GO:0000415) specification of axis polarity(GO:0065001) |
| 0.1 |
0.4 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.1 |
1.4 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 |
0.6 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 |
0.4 |
GO:0071649 |
negative regulation of mitochondrial fusion(GO:0010637) regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.1 |
0.1 |
GO:0051036 |
regulation of endosome size(GO:0051036) |
| 0.1 |
0.1 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 |
0.6 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 |
1.1 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.1 |
0.7 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.1 |
0.3 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.1 |
0.7 |
GO:0006189 |
'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 |
0.5 |
GO:0002901 |
mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 |
0.3 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.1 |
1.5 |
GO:1900037 |
regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 |
1.2 |
GO:0030836 |
positive regulation of actin filament depolymerization(GO:0030836) |
| 0.1 |
0.3 |
GO:1903371 |
regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 |
0.8 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.1 |
0.3 |
GO:0098910 |
regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.1 |
0.5 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 |
1.0 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 |
1.4 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) |
| 0.1 |
0.1 |
GO:0032472 |
Golgi calcium ion transport(GO:0032472) |
| 0.1 |
0.5 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.1 |
0.6 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 |
0.1 |
GO:0071169 |
establishment of protein localization to chromatin(GO:0071169) |
| 0.1 |
0.5 |
GO:0046618 |
drug export(GO:0046618) |
| 0.1 |
1.7 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.1 |
0.3 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 |
0.8 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.1 |
0.8 |
GO:0043137 |
DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 |
0.4 |
GO:2000507 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) circadian sleep/wake cycle, REM sleep(GO:0042747) negative regulation of interleukin-6 biosynthetic process(GO:0045409) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) positive regulation of cortisol secretion(GO:0051464) positive regulation of energy homeostasis(GO:2000507) |
| 0.1 |
0.5 |
GO:0072362 |
regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 |
0.4 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.1 |
0.6 |
GO:0070535 |
histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 |
0.5 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.1 |
1.6 |
GO:0090141 |
positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 |
0.5 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.1 |
0.7 |
GO:0038031 |
non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 |
2.0 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.1 |
0.6 |
GO:0021699 |
cerebellar cortex maturation(GO:0021699) |
| 0.1 |
0.4 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.1 |
1.1 |
GO:0046037 |
GMP metabolic process(GO:0046037) |
| 0.1 |
1.0 |
GO:0000055 |
ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 |
0.8 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 |
0.1 |
GO:0060620 |
regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.1 |
0.5 |
GO:0006481 |
C-terminal protein methylation(GO:0006481) |
| 0.1 |
0.2 |
GO:0006154 |
adenosine catabolic process(GO:0006154) |
| 0.1 |
0.4 |
GO:0048382 |
neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) mesendoderm development(GO:0048382) |
| 0.1 |
7.9 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
0.1 |
GO:0009193 |
pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 |
0.5 |
GO:0015959 |
diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.1 |
0.7 |
GO:1902861 |
copper ion import into cell(GO:1902861) |
| 0.1 |
0.6 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.1 |
0.3 |
GO:2000409 |
positive regulation of T cell extravasation(GO:2000409) |
| 0.1 |
1.4 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 |
3.4 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 |
0.4 |
GO:1903265 |
positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 |
3.6 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 |
0.3 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 |
4.6 |
GO:0045761 |
regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 |
0.5 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 |
0.3 |
GO:1900060 |
negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 |
1.0 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.1 |
0.5 |
GO:0070307 |
lens fiber cell development(GO:0070307) |
| 0.1 |
0.9 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.1 |
1.1 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.1 |
0.3 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 |
0.1 |
GO:0072602 |
interleukin-4 secretion(GO:0072602) |
| 0.1 |
0.3 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 |
0.3 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 |
0.2 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
0.6 |
GO:0000963 |
mitochondrial RNA processing(GO:0000963) |
| 0.1 |
5.6 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.1 |
1.5 |
GO:0050667 |
homocysteine metabolic process(GO:0050667) |
| 0.1 |
0.1 |
GO:0008298 |
intracellular mRNA localization(GO:0008298) |
| 0.1 |
0.5 |
GO:0019249 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.1 |
0.4 |
GO:0040038 |
polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 |
7.0 |
GO:0003281 |
ventricular septum development(GO:0003281) |
| 0.1 |
0.6 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 |
1.7 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
0.3 |
GO:0060971 |
embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 |
0.3 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 |
0.4 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.1 |
0.5 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.3 |
GO:0048698 |
negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 |
0.2 |
GO:0002756 |
MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 |
0.3 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.1 |
0.6 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.1 |
0.5 |
GO:0098543 |
detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.1 |
0.1 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 |
0.3 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 |
0.4 |
GO:0045835 |
negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 |
1.2 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.1 |
0.7 |
GO:0030422 |
production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 |
1.6 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.1 |
0.4 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 |
0.7 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 |
0.4 |
GO:0039532 |
negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.1 |
0.4 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.1 |
1.1 |
GO:0006085 |
acetyl-CoA biosynthetic process(GO:0006085) |
| 0.1 |
0.4 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 |
0.3 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 |
0.5 |
GO:0071800 |
podosome assembly(GO:0071800) |
| 0.1 |
0.4 |
GO:0060632 |
regulation of microtubule-based movement(GO:0060632) |
| 0.1 |
0.4 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.1 |
0.5 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 |
0.4 |
GO:1901673 |
regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 |
0.4 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 |
0.5 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.1 |
0.1 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 |
0.4 |
GO:0009146 |
purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.1 |
0.3 |
GO:0045908 |
negative regulation of vasodilation(GO:0045908) |
| 0.1 |
0.9 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.1 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 |
0.4 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 |
1.2 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.1 |
0.3 |
GO:0019255 |
glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 |
0.3 |
GO:0042505 |
tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.1 |
3.6 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.1 |
1.0 |
GO:0045351 |
type I interferon biosynthetic process(GO:0045351) |
| 0.1 |
0.3 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 |
0.4 |
GO:0006659 |
phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 |
1.7 |
GO:0042059 |
negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.1 |
0.2 |
GO:0036515 |
serotonergic neuron axon guidance(GO:0036515) |
| 0.1 |
0.3 |
GO:1901858 |
regulation of mitochondrial DNA metabolic process(GO:1901858) |
| 0.1 |
0.2 |
GO:0060445 |
branching involved in salivary gland morphogenesis(GO:0060445) regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 |
0.7 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 |
0.9 |
GO:0070932 |
histone H3 deacetylation(GO:0070932) |
| 0.1 |
0.7 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 |
0.3 |
GO:0021678 |
third ventricle development(GO:0021678) |
| 0.1 |
0.2 |
GO:0031126 |
snoRNA 3'-end processing(GO:0031126) |
| 0.1 |
0.2 |
GO:0071281 |
cellular response to iron ion(GO:0071281) |
| 0.1 |
0.9 |
GO:0051352 |
negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.1 |
0.3 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 |
0.5 |
GO:0032596 |
protein transport into membrane raft(GO:0032596) |
| 0.1 |
1.7 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.1 |
0.3 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 |
1.3 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.1 |
0.5 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 |
0.2 |
GO:2000819 |
regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 |
0.9 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 |
0.2 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.1 |
0.1 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.1 |
0.2 |
GO:0046391 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 |
0.5 |
GO:0031145 |
anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 |
0.9 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.1 |
3.5 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.1 |
0.2 |
GO:0072600 |
establishment of protein localization to Golgi(GO:0072600) |
| 0.1 |
0.4 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 |
1.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.1 |
0.6 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 |
0.5 |
GO:0016574 |
histone ubiquitination(GO:0016574) |
| 0.1 |
1.4 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 |
0.1 |
GO:0032532 |
regulation of microvillus organization(GO:0032530) regulation of microvillus length(GO:0032532) |
| 0.1 |
0.4 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 |
0.3 |
GO:0051683 |
establishment of Golgi localization(GO:0051683) |
| 0.1 |
0.2 |
GO:0042275 |
error-free postreplication DNA repair(GO:0042275) |
| 0.1 |
0.5 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
0.6 |
GO:0031998 |
regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 |
0.1 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.1 |
1.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 |
0.1 |
GO:0061724 |
lipophagy(GO:0061724) |
| 0.1 |
0.1 |
GO:1902953 |
positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 |
1.7 |
GO:0030811 |
regulation of glycolytic process(GO:0006110) regulation of nucleotide catabolic process(GO:0030811) |
| 0.1 |
2.8 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 |
0.4 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.1 |
0.4 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 |
0.2 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.1 |
2.1 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.1 |
0.7 |
GO:0090261 |
positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 |
1.1 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 |
0.4 |
GO:0003431 |
growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 |
1.3 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 |
1.0 |
GO:0008088 |
axo-dendritic transport(GO:0008088) |
| 0.1 |
0.1 |
GO:0071865 |
regulation of apoptotic process in bone marrow(GO:0071865) negative regulation of apoptotic process in bone marrow(GO:0071866) |
| 0.1 |
0.2 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.1 |
0.3 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 |
0.2 |
GO:0072356 |
chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 |
0.3 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.1 |
1.4 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 |
0.3 |
GO:0033030 |
negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 |
0.1 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.9 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.1 |
0.1 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) |
| 0.1 |
0.6 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 |
2.4 |
GO:0060612 |
adipose tissue development(GO:0060612) |
| 0.1 |
0.5 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.1 |
0.2 |
GO:0031627 |
telomeric loop formation(GO:0031627) |
| 0.1 |
0.3 |
GO:0003011 |
involuntary skeletal muscle contraction(GO:0003011) |
| 0.1 |
0.3 |
GO:0071639 |
positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.1 |
0.2 |
GO:0030208 |
dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 |
0.7 |
GO:0071549 |
response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 |
0.2 |
GO:1900623 |
positive regulation of keratinocyte proliferation(GO:0010838) endocardial cushion to mesenchymal transition(GO:0090500) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 |
1.3 |
GO:0006693 |
prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 |
0.3 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 |
0.2 |
GO:0045876 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 |
1.0 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.1 |
0.3 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.1 |
0.5 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.1 |
0.2 |
GO:0036216 |
response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 |
0.8 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.1 |
0.3 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
0.2 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.1 |
2.2 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 |
0.1 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.1 |
4.7 |
GO:1903955 |
positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 |
0.5 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 |
2.3 |
GO:0007091 |
metaphase/anaphase transition of mitotic cell cycle(GO:0007091) metaphase/anaphase transition of cell cycle(GO:0044784) |
| 0.1 |
0.7 |
GO:0003094 |
glomerular filtration(GO:0003094) |
| 0.1 |
0.2 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.1 |
0.1 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.1 |
0.4 |
GO:1901798 |
positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 |
5.4 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.1 |
0.4 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.1 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 |
0.2 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 |
0.2 |
GO:0036509 |
trimming of terminal mannose on B branch(GO:0036509) |
| 0.1 |
0.5 |
GO:0043313 |
regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.1 |
0.6 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.1 |
0.8 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.1 |
1.3 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 |
0.2 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 |
0.1 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.1 |
1.3 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 |
0.2 |
GO:0051643 |
endoplasmic reticulum localization(GO:0051643) |
| 0.1 |
0.2 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 |
0.2 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.1 |
0.3 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.1 |
0.7 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.1 |
0.2 |
GO:2000370 |
positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.1 |
0.8 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 |
0.2 |
GO:0072365 |
regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 |
0.5 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 |
0.2 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.1 |
0.4 |
GO:0046716 |
muscle cell cellular homeostasis(GO:0046716) |
| 0.1 |
0.2 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.1 |
0.1 |
GO:0070103 |
regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.1 |
0.1 |
GO:0048239 |
negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 |
1.7 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.1 |
0.2 |
GO:0090153 |
regulation of sphingolipid biosynthetic process(GO:0090153) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.1 |
0.4 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.1 |
0.4 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 |
0.5 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.1 |
0.3 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.1 |
0.2 |
GO:0033182 |
regulation of histone ubiquitination(GO:0033182) |
| 0.1 |
0.8 |
GO:2000369 |
regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 |
1.1 |
GO:0006054 |
N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 |
1.1 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 |
1.6 |
GO:2000036 |
regulation of stem cell population maintenance(GO:2000036) |
| 0.1 |
0.7 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.1 |
0.2 |
GO:0061009 |
common bile duct development(GO:0061009) |
| 0.1 |
0.5 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 |
0.2 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.1 |
0.3 |
GO:0046292 |
formaldehyde metabolic process(GO:0046292) |
| 0.1 |
0.2 |
GO:0010626 |
negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 |
0.2 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 |
0.2 |
GO:0033046 |
negative regulation of sister chromatid segregation(GO:0033046) negative regulation of mitotic sister chromatid segregation(GO:0033048) negative regulation of chromosome segregation(GO:0051985) |
| 0.1 |
0.3 |
GO:0031116 |
positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 |
0.2 |
GO:0071475 |
cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 |
0.7 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.5 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
0.5 |
GO:0061154 |
endothelial tube morphogenesis(GO:0061154) |
| 0.0 |
0.4 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 |
0.1 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.2 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 |
0.2 |
GO:0019348 |
polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 |
0.1 |
GO:0051193 |
regulation of cofactor metabolic process(GO:0051193) |
| 0.0 |
0.9 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.2 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.1 |
GO:1900368 |
regulation of RNA interference(GO:1900368) |
| 0.0 |
0.4 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.0 |
0.4 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.5 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.1 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.4 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.2 |
GO:0090503 |
RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 |
0.2 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.3 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.4 |
GO:0090308 |
regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 |
0.2 |
GO:1904251 |
regulation of bile acid metabolic process(GO:1904251) |
| 0.0 |
2.6 |
GO:0006413 |
translational initiation(GO:0006413) |
| 0.0 |
0.2 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 |
0.3 |
GO:0036066 |
protein O-linked fucosylation(GO:0036066) |
| 0.0 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.8 |
GO:0006182 |
cGMP biosynthetic process(GO:0006182) |
| 0.0 |
2.0 |
GO:0060395 |
SMAD protein signal transduction(GO:0060395) |
| 0.0 |
0.1 |
GO:1903332 |
regulation of protein folding(GO:1903332) |
| 0.0 |
1.9 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.3 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
0.1 |
GO:0052428 |
modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 |
0.6 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 |
0.1 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.0 |
0.1 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 |
0.4 |
GO:0031069 |
hair follicle morphogenesis(GO:0031069) |
| 0.0 |
0.8 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 |
0.1 |
GO:1901978 |
positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 |
0.2 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.0 |
0.2 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.0 |
0.3 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.0 |
GO:0070601 |
centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 |
0.2 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.0 |
0.2 |
GO:1902897 |
regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 |
0.1 |
GO:0097011 |
cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 |
0.1 |
GO:0032570 |
response to progesterone(GO:0032570) |
| 0.0 |
0.6 |
GO:0019730 |
antimicrobial humoral response(GO:0019730) |
| 0.0 |
0.2 |
GO:0097324 |
melanocyte migration(GO:0097324) |
| 0.0 |
0.3 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.0 |
0.2 |
GO:0010992 |
ubiquitin homeostasis(GO:0010992) |
| 0.0 |
0.3 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 |
0.2 |
GO:0000972 |
transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 |
0.3 |
GO:0006878 |
cellular copper ion homeostasis(GO:0006878) |
| 0.0 |
0.5 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.0 |
0.3 |
GO:0007189 |
adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 |
0.1 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 |
0.3 |
GO:0086103 |
G-protein coupled receptor signaling pathway involved in heart process(GO:0086103) |
| 0.0 |
0.1 |
GO:1903799 |
negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 |
0.1 |
GO:0001510 |
RNA methylation(GO:0001510) |
| 0.0 |
0.2 |
GO:0042126 |
nitrate metabolic process(GO:0042126) |
| 0.0 |
0.4 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.0 |
0.4 |
GO:0006383 |
transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 |
0.2 |
GO:0016446 |
somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.7 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.2 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 |
1.0 |
GO:0043038 |
tRNA aminoacylation for protein translation(GO:0006418) amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.0 |
0.1 |
GO:0006451 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.6 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.0 |
0.2 |
GO:0010747 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 |
0.1 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 |
0.6 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.0 |
0.2 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
0.5 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 |
0.2 |
GO:0006353 |
DNA-templated transcription, termination(GO:0006353) |
| 0.0 |
0.3 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.0 |
0.1 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 |
0.2 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 |
0.1 |
GO:0070933 |
histone H4 deacetylation(GO:0070933) |
| 0.0 |
0.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.4 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.1 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 |
0.5 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.4 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.4 |
GO:0006513 |
protein monoubiquitination(GO:0006513) |
| 0.0 |
0.3 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.0 |
0.3 |
GO:0051353 |
positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 |
0.4 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.2 |
GO:0007183 |
SMAD protein complex assembly(GO:0007183) |
| 0.0 |
0.2 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.0 |
0.1 |
GO:0015867 |
ATP transport(GO:0015867) |
| 0.0 |
0.2 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.1 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 |
0.2 |
GO:0010766 |
negative regulation of sodium ion transport(GO:0010766) |
| 0.0 |
0.1 |
GO:0060180 |
female mating behavior(GO:0060180) |
| 0.0 |
0.2 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.0 |
0.5 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.1 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.0 |
0.1 |
GO:1903416 |
regulation of calcidiol 1-monooxygenase activity(GO:0060558) response to glycoside(GO:1903416) |
| 0.0 |
1.0 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.3 |
GO:0050885 |
neuromuscular process controlling balance(GO:0050885) |
| 0.0 |
1.3 |
GO:0060997 |
dendritic spine morphogenesis(GO:0060997) |
| 0.0 |
0.2 |
GO:0035357 |
peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.0 |
0.5 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 |
0.1 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 |
0.2 |
GO:0033146 |
regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 |
0.6 |
GO:0006893 |
Golgi to plasma membrane transport(GO:0006893) |
| 0.0 |
2.9 |
GO:0071805 |
potassium ion transmembrane transport(GO:0071805) |
| 0.0 |
0.2 |
GO:0071158 |
positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 |
0.1 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.0 |
0.1 |
GO:0044321 |
cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 |
0.5 |
GO:0040019 |
positive regulation of embryonic development(GO:0040019) |
| 0.0 |
0.1 |
GO:0051561 |
positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 |
0.2 |
GO:0060547 |
negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 |
2.0 |
GO:0007623 |
circadian rhythm(GO:0007623) |
| 0.0 |
0.2 |
GO:0051646 |
mitochondrion localization(GO:0051646) |
| 0.0 |
0.2 |
GO:1903624 |
regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.0 |
0.4 |
GO:0008214 |
protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 |
0.1 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.0 |
0.3 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.0 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
0.1 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.5 |
GO:0051180 |
vitamin transport(GO:0051180) |
| 0.0 |
0.4 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.3 |
GO:0060117 |
auditory receptor cell development(GO:0060117) |
| 0.0 |
0.1 |
GO:0061668 |
mitochondrial ribosome assembly(GO:0061668) |
| 0.0 |
0.1 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.0 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.8 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 |
0.2 |
GO:2000258 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 |
0.5 |
GO:0045880 |
positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 |
0.1 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 |
0.3 |
GO:0033006 |
regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 |
0.1 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.0 |
0.2 |
GO:0035066 |
positive regulation of histone acetylation(GO:0035066) |
| 0.0 |
0.1 |
GO:0046909 |
intermembrane transport(GO:0046909) |
| 0.0 |
0.3 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.2 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.1 |
GO:0060712 |
spongiotrophoblast layer development(GO:0060712) |
| 0.0 |
0.4 |
GO:0006368 |
transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 |
0.2 |
GO:0031498 |
chromatin disassembly(GO:0031498) |
| 0.0 |
0.1 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.0 |
0.0 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.4 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 |
0.7 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.3 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) |
| 0.0 |
0.3 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.1 |
GO:0051299 |
centrosome separation(GO:0051299) |
| 0.0 |
0.1 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.4 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.0 |
0.1 |
GO:0044351 |
macropinocytosis(GO:0044351) |
| 0.0 |
0.0 |
GO:0016445 |
somatic diversification of immunoglobulins(GO:0016445) |
| 0.0 |
0.1 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 |
0.1 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.0 |
0.2 |
GO:0045669 |
positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 |
0.1 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 |
0.2 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.0 |
0.8 |
GO:0006073 |
glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 |
0.1 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 |
0.1 |
GO:0051533 |
positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 |
0.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.0 |
GO:1902683 |
regulation of protein localization to synapse(GO:1902473) regulation of receptor localization to synapse(GO:1902683) excitatory synapse assembly(GO:1904861) |
| 0.0 |
0.0 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.0 |
0.1 |
GO:0003170 |
heart valve development(GO:0003170) |
| 0.0 |
0.1 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 |
0.2 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.4 |
GO:0048008 |
platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 |
0.0 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 |
0.4 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.2 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.0 |
0.1 |
GO:1902287 |
semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 |
0.1 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.1 |
GO:0060768 |
epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.1 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.0 |
GO:0009838 |
abscission(GO:0009838) |
| 0.0 |
0.1 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.0 |
1.2 |
GO:0048675 |
axon extension(GO:0048675) |
| 0.0 |
0.3 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.6 |
GO:0019233 |
sensory perception of pain(GO:0019233) |
| 0.0 |
0.3 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.1 |
GO:0090045 |
positive regulation of deacetylase activity(GO:0090045) |
| 0.0 |
0.2 |
GO:0001829 |
trophectodermal cell differentiation(GO:0001829) |
| 0.0 |
0.6 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:0031573 |
intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 |
0.1 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 |
0.1 |
GO:0050884 |
neuromuscular process controlling posture(GO:0050884) |
| 0.0 |
0.1 |
GO:0030299 |
intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 |
0.2 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 |
0.1 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |