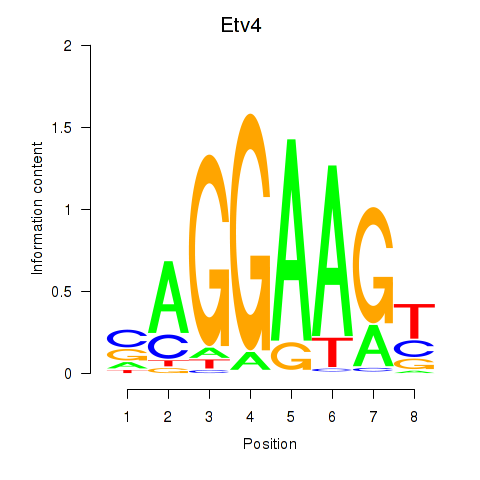
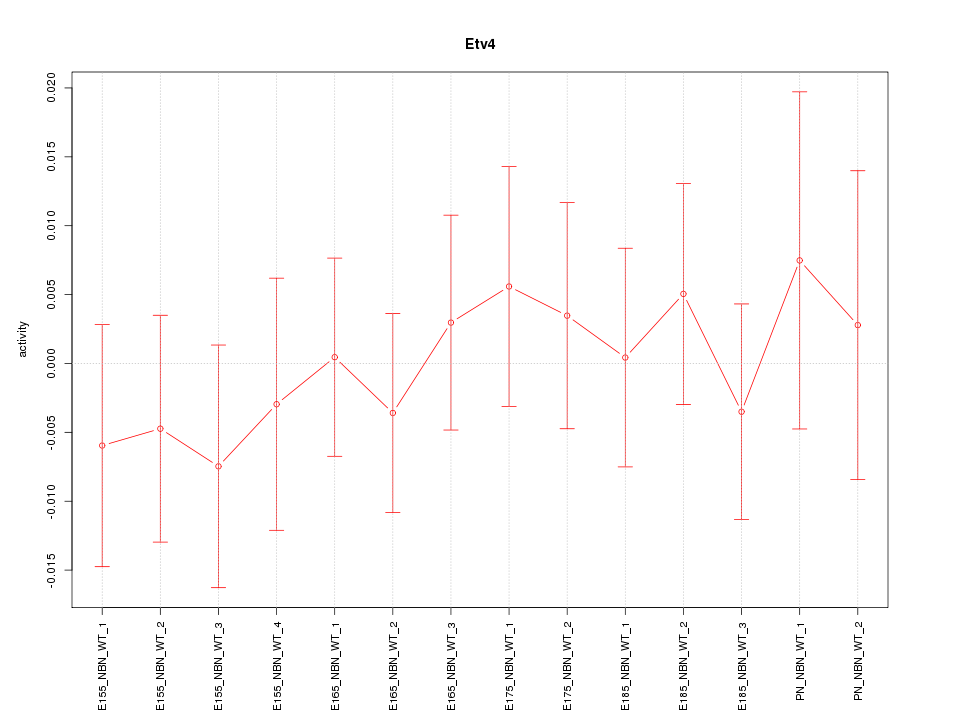
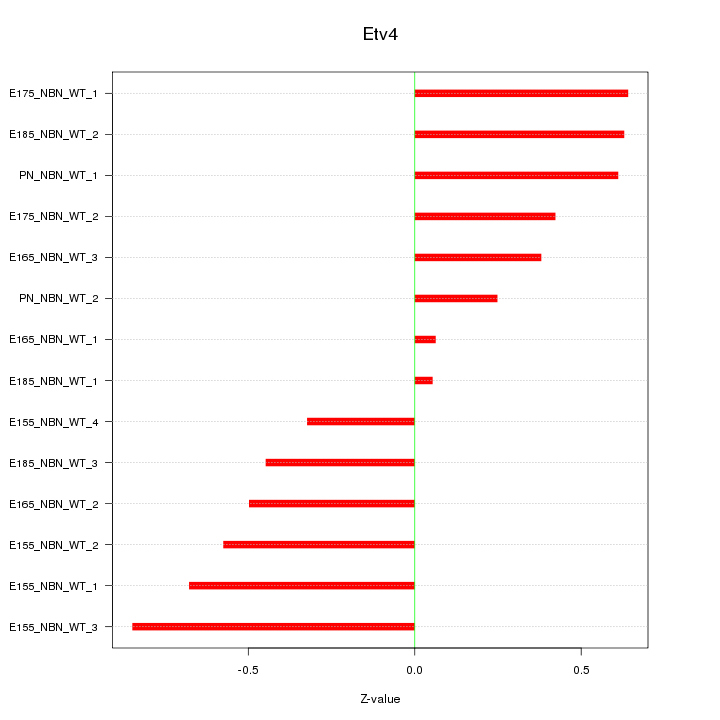
| 0.3 |
0.3 |
GO:0002432 |
granuloma formation(GO:0002432) |
| 0.3 |
0.9 |
GO:0090187 |
zymogen granule exocytosis(GO:0070625) positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.2 |
1.1 |
GO:2001204 |
regulation of osteoclast development(GO:2001204) |
| 0.2 |
0.7 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.2 |
0.7 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 |
0.6 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 |
1.3 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
0.4 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.1 |
0.4 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 |
0.3 |
GO:1902566 |
regulation of eosinophil activation(GO:1902566) |
| 0.1 |
0.4 |
GO:2001187 |
positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.1 |
0.4 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 |
0.3 |
GO:0050859 |
negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 |
1.6 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.1 |
0.6 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 |
0.2 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.1 |
0.3 |
GO:0001803 |
antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.1 |
0.5 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 |
0.2 |
GO:0003032 |
detection of oxygen(GO:0003032) |
| 0.1 |
0.8 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.1 |
0.4 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 |
0.3 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.1 |
0.1 |
GO:0032701 |
negative regulation of interleukin-18 production(GO:0032701) |
| 0.1 |
0.7 |
GO:0045651 |
positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 |
0.2 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 |
0.2 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 |
0.1 |
GO:1902455 |
negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 |
0.2 |
GO:0097168 |
condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 |
0.2 |
GO:0030208 |
dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 |
0.3 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 |
0.3 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.1 |
0.3 |
GO:1900017 |
positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 |
0.2 |
GO:0019043 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) establishment of viral latency(GO:0019043) |
| 0.0 |
0.2 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 |
0.9 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |
| 0.0 |
0.2 |
GO:0098915 |
membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.0 |
0.8 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.3 |
GO:0003065 |
positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 |
0.3 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.0 |
0.1 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.0 |
0.2 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 |
0.1 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.0 |
0.2 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 |
0.3 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 |
0.0 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 |
0.2 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.0 |
0.2 |
GO:0071105 |
response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.0 |
0.3 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 |
0.2 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 |
0.1 |
GO:0090168 |
Golgi reassembly(GO:0090168) regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 |
0.6 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
0.2 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.1 |
GO:0007521 |
muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 |
0.2 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 |
0.1 |
GO:0031052 |
programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.0 |
0.1 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.0 |
0.2 |
GO:0010835 |
regulation of protein ADP-ribosylation(GO:0010835) negative regulation of protein glycosylation(GO:0060051) |
| 0.0 |
0.4 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.4 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.1 |
GO:0035696 |
monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 |
0.1 |
GO:2000642 |
negative regulation of vacuolar transport(GO:1903336) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 |
0.1 |
GO:2000812 |
regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 |
0.1 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
0.2 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 |
0.4 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.2 |
GO:2001054 |
negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 |
0.1 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
0.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.1 |
GO:0048007 |
antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 |
0.1 |
GO:0036118 |
hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 |
0.1 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 |
0.2 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.2 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.1 |
GO:0090365 |
regulation of mRNA modification(GO:0090365) |
| 0.0 |
0.5 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.2 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 |
0.3 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.0 |
0.5 |
GO:0034315 |
regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 |
0.6 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.1 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 |
0.2 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 |
0.5 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.1 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 |
0.1 |
GO:0010730 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 |
0.1 |
GO:0021905 |
pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) |
| 0.0 |
0.1 |
GO:0033278 |
cell proliferation in midbrain(GO:0033278) |
| 0.0 |
0.1 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 |
0.1 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.0 |
0.0 |
GO:2000425 |
regulation of apoptotic cell clearance(GO:2000425) |
| 0.0 |
0.1 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 |
0.7 |
GO:0071353 |
response to interleukin-4(GO:0070670) cellular response to interleukin-4(GO:0071353) |
| 0.0 |
0.3 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.2 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 |
0.1 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.1 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 |
0.5 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
0.2 |
GO:0017145 |
stem cell division(GO:0017145) |
| 0.0 |
0.6 |
GO:0030488 |
tRNA methylation(GO:0030488) |
| 0.0 |
0.2 |
GO:0071285 |
cellular response to lithium ion(GO:0071285) |
| 0.0 |
0.2 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.0 |
0.1 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 |
0.1 |
GO:0045346 |
regulation of MHC class II biosynthetic process(GO:0045346) negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 |
0.1 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 |
0.2 |
GO:0006273 |
lagging strand elongation(GO:0006273) |
| 0.0 |
0.2 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 |
0.2 |
GO:0032096 |
negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 |
0.1 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.0 |
0.1 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 |
0.1 |
GO:2000002 |
negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 |
0.1 |
GO:0009212 |
dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 |
0.7 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.2 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.0 |
0.0 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.1 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 |
0.1 |
GO:0070863 |
positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 |
0.1 |
GO:0090220 |
meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 |
0.3 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.0 |
0.3 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.0 |
0.1 |
GO:0006451 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.1 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.0 |
0.3 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.0 |
0.1 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 |
0.1 |
GO:1900110 |
negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 |
0.1 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.1 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 |
0.6 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.1 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.1 |
GO:0032211 |
negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 |
0.1 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 |
0.4 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.5 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.0 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 |
0.1 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 |
0.2 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 |
0.1 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.0 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.0 |
0.5 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 |
0.1 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.0 |
0.1 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.0 |
0.1 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.0 |
0.2 |
GO:0010039 |
response to iron ion(GO:0010039) |
| 0.0 |
0.3 |
GO:0002718 |
regulation of cytokine production involved in immune response(GO:0002718) |
| 0.0 |
0.1 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 |
0.0 |
GO:0010989 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 |
0.1 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.1 |
GO:0044144 |
regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 |
0.1 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.1 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.2 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.1 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 |
0.1 |
GO:1901725 |
regulation of histone deacetylase activity(GO:1901725) |
| 0.0 |
0.3 |
GO:0018279 |
protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.0 |
GO:0045472 |
response to ether(GO:0045472) |
| 0.0 |
0.1 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 |
0.1 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 |
0.1 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.1 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.1 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.0 |
0.1 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.1 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.0 |
0.0 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.0 |
0.2 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |