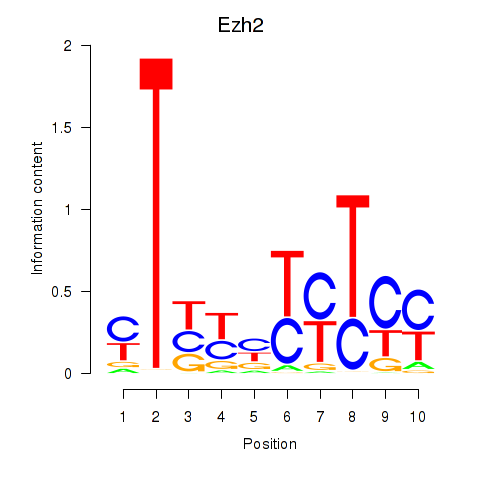
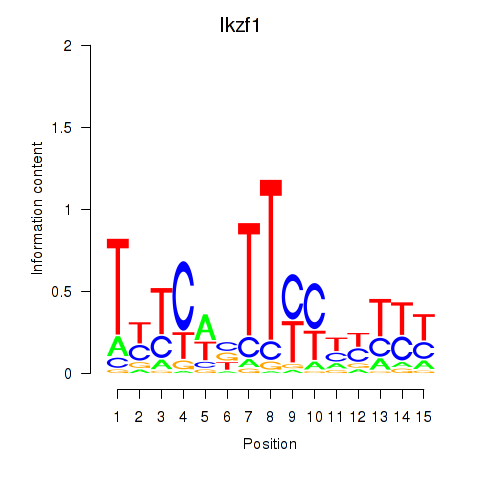
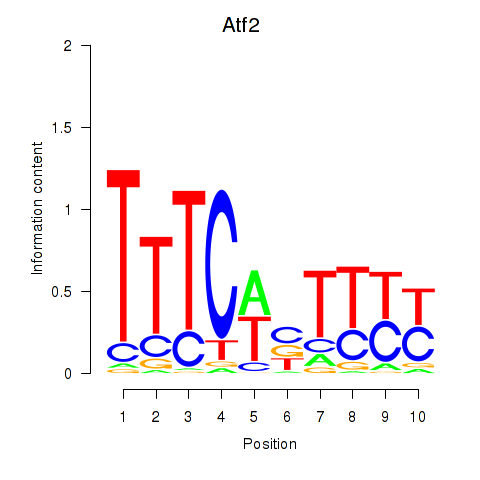
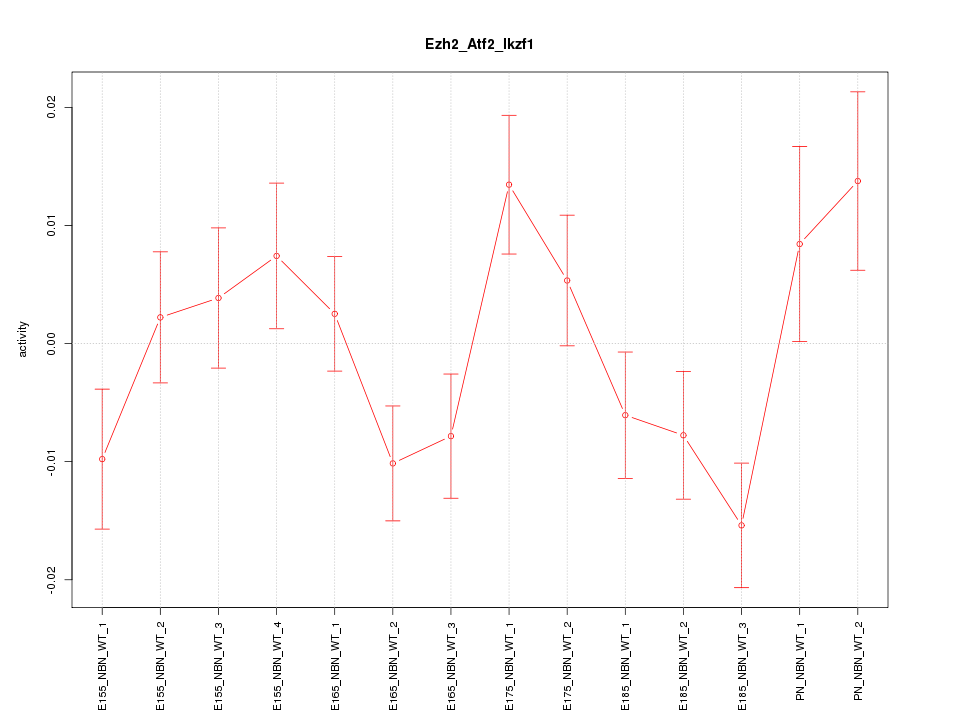
| 2.5 |
7.5 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.1 |
3.3 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 1.0 |
4.1 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.7 |
3.5 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.7 |
2.1 |
GO:0060067 |
cervix development(GO:0060067) |
| 0.7 |
2.0 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.3 |
0.7 |
GO:0034372 |
very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.3 |
0.9 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.3 |
1.2 |
GO:1903984 |
positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.3 |
1.4 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 0.3 |
1.3 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.3 |
1.0 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.2 |
1.0 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.2 |
0.4 |
GO:2000790 |
regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.2 |
1.0 |
GO:0021856 |
cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.2 |
0.8 |
GO:0055099 |
regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 0.2 |
0.6 |
GO:0007521 |
muscle cell fate determination(GO:0007521) |
| 0.2 |
1.3 |
GO:0090188 |
negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.2 |
0.6 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.2 |
0.5 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.2 |
0.8 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.2 |
0.5 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.2 |
0.6 |
GO:0098912 |
membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.1 |
0.3 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.1 |
5.3 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.7 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 |
0.4 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.1 |
1.4 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.4 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) |
| 0.1 |
0.7 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.4 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 |
0.4 |
GO:0021553 |
establishment of blood-nerve barrier(GO:0008065) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) olfactory nerve development(GO:0021553) |
| 0.1 |
0.5 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 |
0.5 |
GO:0032901 |
positive regulation of neurotrophin production(GO:0032901) |
| 0.1 |
0.8 |
GO:0019336 |
phenol-containing compound catabolic process(GO:0019336) |
| 0.1 |
0.9 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 |
0.2 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.1 |
0.5 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.1 |
0.5 |
GO:2000858 |
mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) positive regulation of mineralocorticoid secretion(GO:2000857) regulation of aldosterone secretion(GO:2000858) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 |
0.3 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 |
0.8 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.1 |
4.5 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.1 |
0.3 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.1 |
0.4 |
GO:1903251 |
multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 |
0.4 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 |
0.4 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.1 |
0.2 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.1 |
1.6 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
0.9 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 |
1.4 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
0.4 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.1 |
0.3 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.1 |
0.7 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.1 |
0.3 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.1 |
0.3 |
GO:1903800 |
astrocyte activation(GO:0048143) positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 |
1.2 |
GO:0006309 |
apoptotic DNA fragmentation(GO:0006309) |
| 0.1 |
0.3 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
0.5 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.1 |
0.6 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
0.2 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 0.1 |
0.7 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
1.1 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
0.2 |
GO:0045938 |
positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 |
0.1 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 |
3.5 |
GO:0001919 |
regulation of receptor recycling(GO:0001919) |
| 0.1 |
0.7 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.4 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 |
0.4 |
GO:0070072 |
vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 |
1.8 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
0.6 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.1 |
0.5 |
GO:1900116 |
sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 |
0.2 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.1 |
0.1 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) |
| 0.1 |
0.1 |
GO:0045981 |
positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.1 |
0.2 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 |
0.5 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.1 |
0.4 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.1 |
0.3 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 |
0.4 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.1 |
0.3 |
GO:1904996 |
positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 |
0.2 |
GO:1904956 |
regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 |
0.2 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 |
0.3 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 |
0.2 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.1 |
1.2 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.1 |
0.2 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.1 |
0.2 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 |
0.3 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 |
0.2 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.1 |
0.2 |
GO:0048682 |
axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 |
0.6 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.1 |
0.2 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.1 |
0.1 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.1 |
0.4 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 |
1.0 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 |
0.2 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.1 |
0.3 |
GO:0032237 |
activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 |
0.2 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 |
0.3 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
0.2 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 0.1 |
0.2 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.1 |
0.3 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 |
0.2 |
GO:0000821 |
regulation of arginine metabolic process(GO:0000821) |
| 0.1 |
0.1 |
GO:0034380 |
high-density lipoprotein particle assembly(GO:0034380) |
| 0.1 |
0.5 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.1 |
0.3 |
GO:0060340 |
positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 |
0.1 |
GO:1903279 |
regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 |
0.2 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.1 |
0.2 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.1 |
3.3 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.1 |
0.2 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 |
0.2 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 |
0.2 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 |
0.2 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 |
0.2 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.5 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.2 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 |
0.3 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.1 |
GO:1903288 |
regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 |
0.2 |
GO:2000325 |
regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 |
0.1 |
GO:1902608 |
regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 |
0.6 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.2 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 0.0 |
0.0 |
GO:0002371 |
dendritic cell cytokine production(GO:0002371) regulation of dendritic cell cytokine production(GO:0002730) regulation of interleukin-18 production(GO:0032661) negative regulation of interleukin-18 production(GO:0032701) |
| 0.0 |
0.0 |
GO:0010593 |
negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 |
0.2 |
GO:0051890 |
regulation of cardioblast differentiation(GO:0051890) positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 |
0.6 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 |
0.3 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 |
0.1 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 |
0.4 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.0 |
0.3 |
GO:0031508 |
pericentric heterochromatin assembly(GO:0031508) |
| 0.0 |
0.1 |
GO:0097105 |
presynaptic membrane assembly(GO:0097105) |
| 0.0 |
0.0 |
GO:0032829 |
regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.0 |
0.2 |
GO:0001542 |
ovulation from ovarian follicle(GO:0001542) positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 |
0.4 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 |
0.0 |
GO:0036336 |
dendritic cell chemotaxis(GO:0002407) myeloid dendritic cell chemotaxis(GO:0002408) dendritic cell migration(GO:0036336) |
| 0.0 |
0.5 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.0 |
0.4 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 |
0.2 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 |
0.6 |
GO:0090129 |
positive regulation of synapse maturation(GO:0090129) |
| 0.0 |
4.2 |
GO:1900006 |
positive regulation of dendrite development(GO:1900006) |
| 0.0 |
0.3 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.1 |
GO:0002765 |
immune response-inhibiting signal transduction(GO:0002765) |
| 0.0 |
0.2 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 |
0.3 |
GO:0032693 |
negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 |
0.1 |
GO:0021837 |
motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.0 |
0.1 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 |
0.1 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 |
0.1 |
GO:1904192 |
cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 |
0.2 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.1 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.0 |
0.2 |
GO:0042447 |
hormone catabolic process(GO:0042447) |
| 0.0 |
0.1 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.0 |
0.4 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.0 |
0.4 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
3.8 |
GO:0045727 |
positive regulation of translation(GO:0045727) |
| 0.0 |
0.4 |
GO:0043116 |
negative regulation of vascular permeability(GO:0043116) |
| 0.0 |
0.4 |
GO:1901844 |
regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 |
0.4 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 |
0.2 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 |
0.6 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
1.4 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.4 |
GO:0060049 |
regulation of protein glycosylation(GO:0060049) |
| 0.0 |
0.2 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.0 |
GO:0033605 |
positive regulation of catecholamine secretion(GO:0033605) |
| 0.0 |
0.6 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.0 |
0.1 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.0 |
0.3 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.1 |
GO:0006824 |
cobalt ion transport(GO:0006824) |
| 0.0 |
0.1 |
GO:0097475 |
motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 |
0.3 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.2 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 |
0.6 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.0 |
0.1 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.0 |
0.2 |
GO:0002674 |
negative regulation of acute inflammatory response(GO:0002674) |
| 0.0 |
0.2 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.0 |
0.2 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 |
0.4 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 |
0.2 |
GO:0090160 |
Golgi to lysosome transport(GO:0090160) |
| 0.0 |
0.1 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.0 |
0.5 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
0.2 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.1 |
GO:0036376 |
sodium ion export from cell(GO:0036376) |
| 0.0 |
0.8 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
0.2 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.1 |
GO:0031652 |
positive regulation of heat generation(GO:0031652) |
| 0.0 |
0.8 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.4 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.0 |
0.1 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.0 |
0.2 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) dendritic spine maintenance(GO:0097062) |
| 0.0 |
0.3 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.4 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.3 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.1 |
GO:2000418 |
positive regulation of eosinophil migration(GO:2000418) |
| 0.0 |
0.3 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.0 |
0.2 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 |
0.1 |
GO:0046880 |
regulation of gonadotropin secretion(GO:0032276) regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 |
0.5 |
GO:0051974 |
negative regulation of telomerase activity(GO:0051974) |
| 0.0 |
0.3 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.0 |
0.3 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 |
2.1 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.1 |
GO:1904754 |
positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.0 |
0.1 |
GO:0003360 |
brainstem development(GO:0003360) |
| 0.0 |
0.2 |
GO:0070102 |
interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 |
0.3 |
GO:0006273 |
lagging strand elongation(GO:0006273) |
| 0.0 |
0.4 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 |
0.0 |
GO:0002524 |
hypersensitivity(GO:0002524) |
| 0.0 |
0.5 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.0 |
0.4 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 |
0.2 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 |
1.0 |
GO:0007628 |
adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 |
0.1 |
GO:0006547 |
histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.0 |
0.3 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 |
0.2 |
GO:0036120 |
response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 |
0.1 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 |
0.1 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 |
0.7 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.1 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.0 |
0.4 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.0 |
GO:0035910 |
ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 |
0.2 |
GO:0032793 |
positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 |
0.1 |
GO:0046601 |
positive regulation of centriole replication(GO:0046601) |
| 0.0 |
0.2 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.4 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.1 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 |
0.0 |
GO:0071442 |
regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.3 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.4 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.1 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 |
0.2 |
GO:0070633 |
transepithelial transport(GO:0070633) |
| 0.0 |
0.1 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 |
0.4 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.2 |
GO:1900120 |
regulation of receptor binding(GO:1900120) |
| 0.0 |
0.5 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.5 |
GO:0048814 |
regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 |
0.1 |
GO:1901678 |
iron coordination entity transport(GO:1901678) |
| 0.0 |
0.1 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.0 |
1.4 |
GO:0007631 |
feeding behavior(GO:0007631) |
| 0.0 |
0.1 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 |
0.2 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.0 |
0.2 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
0.0 |
GO:0072071 |
mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) |
| 0.0 |
0.5 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
0.4 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.0 |
GO:1900747 |
negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 |
0.6 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.1 |
GO:0031055 |
chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.1 |
GO:0002693 |
positive regulation of cellular extravasation(GO:0002693) |
| 0.0 |
0.1 |
GO:0043117 |
positive regulation of vascular permeability(GO:0043117) |
| 0.0 |
0.1 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.0 |
0.0 |
GO:0060457 |
negative regulation of digestive system process(GO:0060457) |
| 0.0 |
0.1 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.1 |
GO:0048520 |
positive regulation of behavior(GO:0048520) |
| 0.0 |
0.1 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 |
0.1 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 |
0.0 |
GO:0036112 |
medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 |
0.1 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.0 |
0.1 |
GO:0030208 |
dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 |
0.2 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.0 |
0.1 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.0 |
0.1 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 |
1.2 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:0006285 |
base-excision repair, AP site formation(GO:0006285) |
| 0.0 |
0.2 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
2.2 |
GO:0071805 |
potassium ion transmembrane transport(GO:0071805) |
| 0.0 |
0.0 |
GO:0008594 |
photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 |
0.1 |
GO:0046889 |
positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 |
0.3 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.1 |
GO:0032053 |
ciliary basal body organization(GO:0032053) |
| 0.0 |
0.2 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.1 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.0 |
0.3 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.1 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.0 |
GO:1901386 |
negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 |
0.2 |
GO:0036037 |
CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.1 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 |
0.1 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
0.0 |
GO:0090238 |
positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 |
0.1 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.1 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.1 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 |
0.2 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.1 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.0 |
0.2 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.1 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.1 |
GO:0015867 |
ATP transport(GO:0015867) |
| 0.0 |
0.1 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 |
0.2 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
0.1 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.0 |
0.0 |
GO:0030948 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 |
0.1 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.1 |
GO:0050716 |
regulation of interleukin-1 secretion(GO:0050704) regulation of interleukin-1 beta secretion(GO:0050706) positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 |
0.5 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 |
1.0 |
GO:0007286 |
spermatid development(GO:0007286) |
| 0.0 |
0.1 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
0.1 |
GO:0042126 |
nitrate metabolic process(GO:0042126) |
| 0.0 |
0.1 |
GO:0008300 |
isoprenoid catabolic process(GO:0008300) |
| 0.0 |
0.2 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.0 |
GO:0090192 |
regulation of glomerular mesangial cell proliferation(GO:0072124) regulation of glomerulus development(GO:0090192) |
| 0.0 |
0.4 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.0 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.0 |
0.1 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.1 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.0 |
0.3 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.1 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.1 |
GO:0051900 |
regulation of mitochondrial depolarization(GO:0051900) |
| 0.0 |
0.1 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 |
0.1 |
GO:1901629 |
presynaptic membrane organization(GO:0097090) regulation of presynaptic membrane organization(GO:1901629) |
| 0.0 |
0.1 |
GO:0060330 |
regulation of response to interferon-gamma(GO:0060330) |
| 0.0 |
0.1 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
0.1 |
GO:0080182 |
histone H3-K4 trimethylation(GO:0080182) |
| 0.0 |
0.5 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 |
0.2 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 |
0.1 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.0 |
0.1 |
GO:0043619 |
regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 |
0.1 |
GO:1901162 |
serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 |
0.1 |
GO:0070827 |
chromatin maintenance(GO:0070827) |
| 0.0 |
0.4 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 |
0.4 |
GO:0014037 |
Schwann cell differentiation(GO:0014037) |
| 0.0 |
0.2 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.0 |
0.1 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.1 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.1 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.0 |
GO:0051096 |
regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |