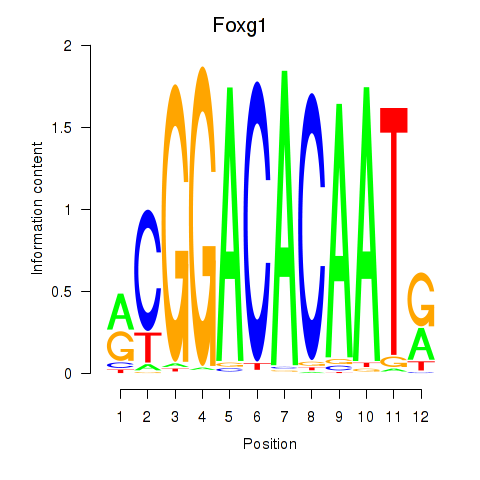
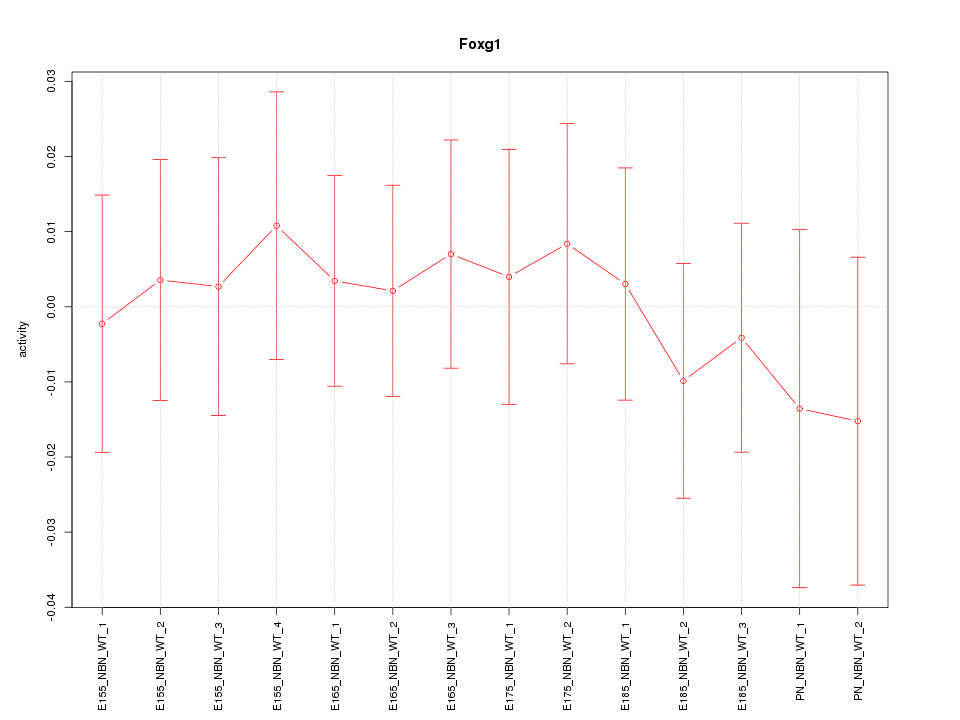
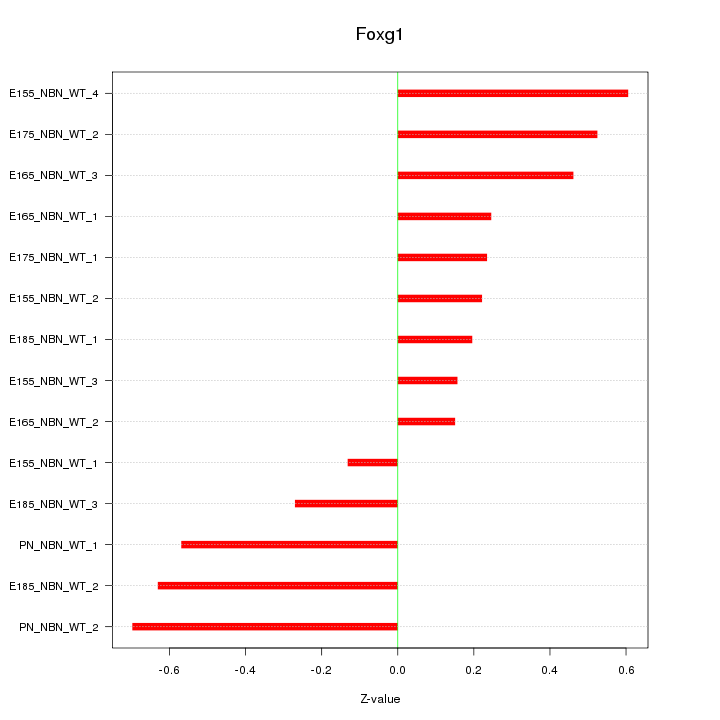
| 0.5 |
2.0 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.3 |
0.9 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.2 |
1.8 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 |
0.8 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 |
0.7 |
GO:0042984 |
amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 |
0.3 |
GO:0071335 |
negative regulation of Schwann cell proliferation(GO:0010626) myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) hair follicle cell proliferation(GO:0071335) |
| 0.1 |
0.1 |
GO:0070346 |
positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 |
0.2 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.0 |
0.2 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.0 |
0.4 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.1 |
GO:2000832 |
negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 |
0.2 |
GO:0036265 |
RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 |
0.2 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.1 |
GO:0070428 |
regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 |
0.5 |
GO:0007289 |
spermatid nucleus differentiation(GO:0007289) |
| 0.0 |
0.2 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.0 |
0.4 |
GO:0071549 |
cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 |
0.3 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 |
0.3 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.1 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.2 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.0 |
0.1 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |