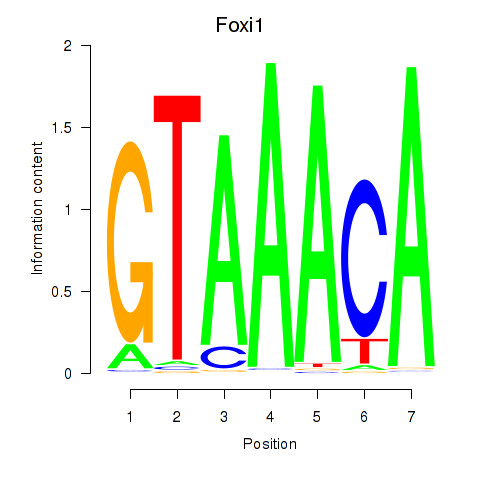
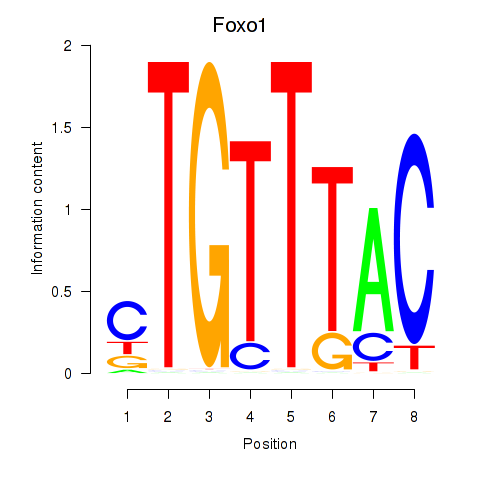
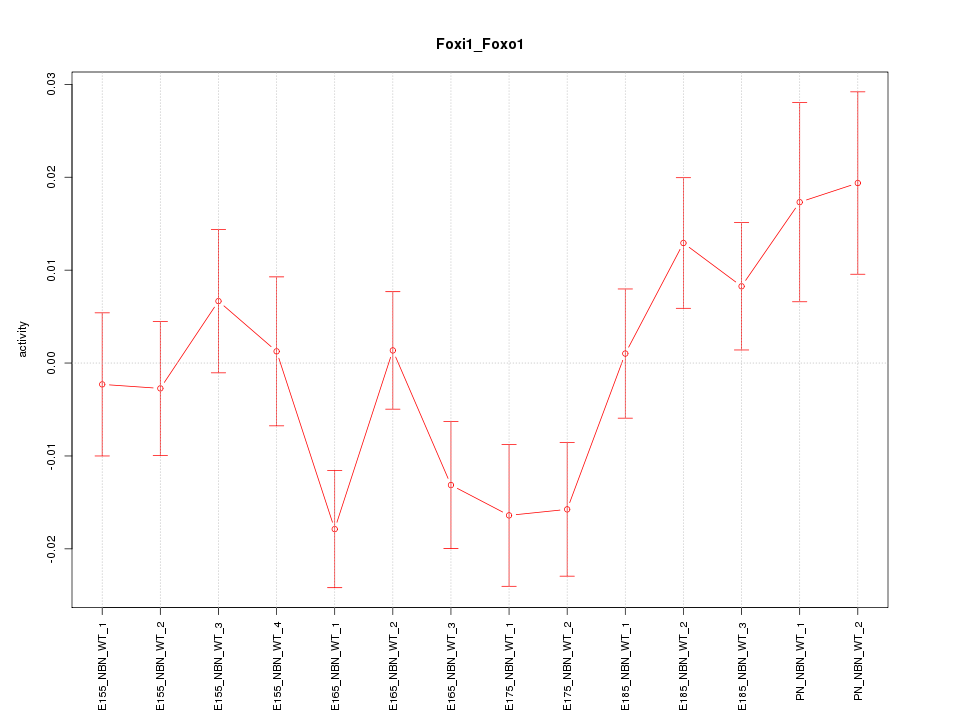
| 1.9 |
9.5 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 1.4 |
4.3 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.8 |
2.4 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.8 |
2.4 |
GO:1901254 |
positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.8 |
7.8 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.8 |
2.3 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.8 |
3.0 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.7 |
2.2 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.7 |
2.1 |
GO:2001139 |
negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 0.7 |
4.7 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.6 |
2.4 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.6 |
1.7 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.5 |
1.6 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.5 |
1.6 |
GO:0072076 |
nephrogenic mesenchyme development(GO:0072076) |
| 0.5 |
2.0 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.5 |
2.0 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.5 |
1.4 |
GO:1902524 |
negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.5 |
1.4 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.5 |
0.5 |
GO:0032769 |
negative regulation of monooxygenase activity(GO:0032769) negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.5 |
1.4 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.4 |
1.3 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.4 |
4.0 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 0.4 |
3.6 |
GO:0021984 |
adenohypophysis development(GO:0021984) |
| 0.4 |
5.5 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.3 |
1.4 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 |
3.8 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.3 |
1.4 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.3 |
1.6 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.3 |
1.0 |
GO:0031990 |
RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.3 |
1.6 |
GO:0031052 |
programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.3 |
1.2 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.3 |
0.6 |
GO:0051464 |
positive regulation of cortisol secretion(GO:0051464) |
| 0.3 |
1.5 |
GO:0034374 |
low-density lipoprotein particle remodeling(GO:0034374) |
| 0.3 |
0.9 |
GO:1902044 |
regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.3 |
1.4 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.3 |
1.7 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 |
3.7 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.3 |
0.8 |
GO:0042908 |
xenobiotic transport(GO:0042908) |
| 0.3 |
0.8 |
GO:0006001 |
fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.3 |
0.5 |
GO:2000504 |
positive regulation of blood vessel remodeling(GO:2000504) |
| 0.3 |
1.8 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 |
1.0 |
GO:1990086 |
lens fiber cell apoptotic process(GO:1990086) |
| 0.3 |
0.8 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.3 |
0.8 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.2 |
0.7 |
GO:1902866 |
regulation of retina development in camera-type eye(GO:1902866) |
| 0.2 |
0.7 |
GO:2000598 |
regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.2 |
0.9 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.2 |
0.9 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.2 |
0.9 |
GO:0051593 |
response to folic acid(GO:0051593) |
| 0.2 |
1.5 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 |
0.9 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 |
0.7 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.2 |
1.3 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 |
0.9 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 |
1.9 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.2 |
0.4 |
GO:0050812 |
regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.2 |
0.6 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 |
1.0 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 |
0.8 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 |
2.0 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.2 |
1.8 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.2 |
0.2 |
GO:0071442 |
regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 |
0.6 |
GO:0003032 |
detection of oxygen(GO:0003032) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.2 |
1.0 |
GO:1990314 |
cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.2 |
1.0 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.2 |
0.6 |
GO:0086069 |
desmosome assembly(GO:0002159) response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.2 |
0.6 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 |
4.5 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.2 |
1.2 |
GO:0071499 |
response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.2 |
2.7 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.2 |
0.9 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.2 |
1.5 |
GO:0002347 |
response to tumor cell(GO:0002347) |
| 0.2 |
0.7 |
GO:2000668 |
dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.2 |
0.9 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.2 |
0.7 |
GO:0097070 |
ductus arteriosus closure(GO:0097070) |
| 0.2 |
1.3 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.2 |
0.7 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.2 |
0.3 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.2 |
0.5 |
GO:0006285 |
base-excision repair, AP site formation(GO:0006285) |
| 0.2 |
0.5 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 |
0.5 |
GO:0060854 |
cell quiescence(GO:0044838) patterning of lymph vessels(GO:0060854) |
| 0.2 |
0.5 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 |
0.5 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.2 |
0.5 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.2 |
0.6 |
GO:1900864 |
mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 |
0.9 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.2 |
1.2 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.2 |
1.5 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.2 |
0.2 |
GO:1905050 |
positive regulation of metallopeptidase activity(GO:1905050) |
| 0.2 |
0.5 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 |
1.0 |
GO:1901162 |
serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 |
0.4 |
GO:0009812 |
flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.1 |
3.7 |
GO:0001893 |
maternal placenta development(GO:0001893) |
| 0.1 |
0.1 |
GO:0048859 |
formation of anatomical boundary(GO:0048859) |
| 0.1 |
0.4 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 |
1.2 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.1 |
3.8 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 |
0.6 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 |
0.6 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.1 |
1.0 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 |
0.6 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.1 |
1.1 |
GO:1903960 |
negative regulation of anion transmembrane transport(GO:1903960) |
| 0.1 |
1.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
0.8 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.3 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 |
0.8 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 |
1.5 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
1.7 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.1 |
2.5 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 |
0.8 |
GO:0045630 |
positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.1 |
1.8 |
GO:0010574 |
regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.1 |
0.6 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.1 |
0.6 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 |
0.5 |
GO:2000675 |
negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 |
0.9 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
0.8 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.1 |
0.4 |
GO:1902071 |
hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 |
0.5 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 |
1.0 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
0.4 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 |
0.2 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.1 |
1.3 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
0.7 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 |
0.4 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.1 |
0.4 |
GO:0071105 |
response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.1 |
0.2 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 |
0.3 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 |
0.3 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.1 |
0.9 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 |
0.4 |
GO:0036006 |
response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.1 |
0.5 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 |
0.6 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.1 |
2.8 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
0.6 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.1 |
1.1 |
GO:0045624 |
positive regulation of T-helper cell differentiation(GO:0045624) |
| 0.1 |
0.4 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 |
0.2 |
GO:2001054 |
negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 |
0.2 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.1 |
0.5 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 |
1.7 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 |
0.7 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.1 |
1.1 |
GO:0060586 |
multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.1 |
0.5 |
GO:0060576 |
intestinal epithelial cell development(GO:0060576) |
| 0.1 |
0.4 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 |
0.2 |
GO:0070427 |
nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 |
0.4 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.1 |
0.6 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 |
1.4 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 |
0.3 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.1 |
0.5 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 |
1.1 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 |
1.4 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 |
0.3 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 |
0.8 |
GO:0035729 |
response to hepatocyte growth factor(GO:0035728) cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.1 |
0.5 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 |
0.6 |
GO:0042095 |
interferon-gamma biosynthetic process(GO:0042095) |
| 0.1 |
0.5 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 |
0.8 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 |
0.9 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
0.6 |
GO:0002643 |
regulation of tolerance induction(GO:0002643) |
| 0.1 |
1.0 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
0.3 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 0.1 |
0.4 |
GO:2000973 |
regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 |
0.3 |
GO:0006665 |
sphingolipid metabolic process(GO:0006665) |
| 0.1 |
0.3 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.1 |
0.5 |
GO:0007128 |
meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.1 |
0.2 |
GO:2001268 |
negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 |
0.2 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 |
0.3 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 |
0.7 |
GO:2000505 |
regulation of energy homeostasis(GO:2000505) |
| 0.1 |
0.4 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
0.4 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 |
0.6 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
0.5 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 |
1.5 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.1 |
0.4 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.3 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 |
0.3 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.1 |
0.3 |
GO:0006545 |
glycine biosynthetic process(GO:0006545) |
| 0.1 |
1.5 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.1 |
0.4 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 |
0.5 |
GO:0009068 |
aspartate family amino acid catabolic process(GO:0009068) |
| 0.1 |
1.0 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 |
1.6 |
GO:0006767 |
water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 |
0.6 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
1.0 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.1 |
0.4 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 |
0.5 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.1 |
0.2 |
GO:0045448 |
regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 |
0.4 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 |
1.2 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.1 |
2.3 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.1 |
0.3 |
GO:0071051 |
polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 |
0.9 |
GO:0043970 |
histone H3-K9 acetylation(GO:0043970) |
| 0.1 |
0.2 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.1 |
0.9 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 |
0.4 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.1 |
0.2 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 |
0.2 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 |
0.9 |
GO:1900746 |
regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.1 |
0.2 |
GO:0002339 |
B cell selection(GO:0002339) B cell negative selection(GO:0002352) response to mycotoxin(GO:0010046) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 |
0.1 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 0.1 |
0.2 |
GO:0044331 |
cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.1 |
0.4 |
GO:0031100 |
organ regeneration(GO:0031100) |
| 0.1 |
0.4 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 |
0.3 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 |
1.0 |
GO:0051150 |
regulation of smooth muscle cell differentiation(GO:0051150) |
| 0.1 |
1.0 |
GO:0019369 |
arachidonic acid metabolic process(GO:0019369) |
| 0.1 |
0.3 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.1 |
0.9 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 |
0.3 |
GO:0006529 |
asparagine biosynthetic process(GO:0006529) |
| 0.1 |
0.2 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 |
0.7 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.1 |
0.1 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.1 |
0.8 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 |
1.7 |
GO:1901661 |
quinone metabolic process(GO:1901661) |
| 0.1 |
0.5 |
GO:0030224 |
monocyte differentiation(GO:0030224) |
| 0.1 |
1.7 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 |
2.0 |
GO:0042168 |
heme metabolic process(GO:0042168) |
| 0.1 |
0.4 |
GO:0001302 |
replicative cell aging(GO:0001302) |
| 0.1 |
1.3 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.1 |
0.2 |
GO:0060455 |
negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 |
0.8 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.1 |
0.2 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 |
0.9 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.1 |
0.2 |
GO:0051541 |
elastin metabolic process(GO:0051541) |
| 0.1 |
0.2 |
GO:0021940 |
positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 |
0.3 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 |
0.3 |
GO:0070234 |
positive regulation of T cell apoptotic process(GO:0070234) |
| 0.1 |
1.5 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 |
0.3 |
GO:0051461 |
corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 |
0.6 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.1 |
0.2 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.1 |
0.3 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 |
0.5 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.1 |
0.2 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 0.1 |
0.2 |
GO:0007525 |
somatic muscle development(GO:0007525) axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 |
1.6 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
0.5 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 |
0.6 |
GO:0032367 |
intracellular cholesterol transport(GO:0032367) |
| 0.1 |
0.2 |
GO:0035022 |
positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 |
0.3 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.8 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.1 |
1.7 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 |
0.2 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 |
0.1 |
GO:0060353 |
cell adhesion molecule production(GO:0060352) regulation of cell adhesion molecule production(GO:0060353) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 |
0.2 |
GO:0006363 |
termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 |
1.5 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.2 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.0 |
1.5 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.0 |
0.1 |
GO:0060406 |
positive regulation of penile erection(GO:0060406) |
| 0.0 |
0.1 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 |
0.2 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 |
1.1 |
GO:0050819 |
negative regulation of coagulation(GO:0050819) |
| 0.0 |
0.7 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
1.3 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.2 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 |
0.0 |
GO:0006532 |
aspartate biosynthetic process(GO:0006532) |
| 0.0 |
0.1 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.0 |
0.5 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
0.2 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.6 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 |
1.3 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.1 |
GO:0002931 |
response to ischemia(GO:0002931) |
| 0.0 |
0.7 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.0 |
0.2 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.0 |
0.4 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.2 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.0 |
1.3 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.4 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.0 |
GO:0060681 |
branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 |
0.0 |
GO:0075136 |
response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) neutrophil mediated killing of bacterium(GO:0070944) response to host(GO:0075136) |
| 0.0 |
1.6 |
GO:0042058 |
regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 |
1.4 |
GO:0046825 |
regulation of protein export from nucleus(GO:0046825) |
| 0.0 |
0.7 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 |
0.1 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 |
0.4 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.0 |
0.2 |
GO:0035360 |
positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 |
0.2 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.0 |
0.2 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 |
0.1 |
GO:0002677 |
negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 |
0.1 |
GO:0010803 |
regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 |
0.8 |
GO:1904355 |
positive regulation of telomere capping(GO:1904355) |
| 0.0 |
1.0 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.2 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.8 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
1.1 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.0 |
0.3 |
GO:1900364 |
negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 |
0.5 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.0 |
0.1 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 |
0.1 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 |
0.1 |
GO:1902255 |
positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 |
1.4 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
2.4 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.6 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.0 |
1.0 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.5 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.0 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 |
0.2 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.3 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.1 |
GO:1901386 |
negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 |
0.1 |
GO:0042428 |
serotonin metabolic process(GO:0042428) |
| 0.0 |
0.2 |
GO:0033210 |
leptin-mediated signaling pathway(GO:0033210) |
| 0.0 |
1.3 |
GO:0086003 |
cardiac muscle cell contraction(GO:0086003) |
| 0.0 |
0.3 |
GO:0071549 |
response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 |
0.6 |
GO:0006182 |
cGMP biosynthetic process(GO:0006182) |
| 0.0 |
0.1 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.0 |
0.5 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.1 |
GO:0003057 |
regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 |
0.1 |
GO:0036451 |
cap mRNA methylation(GO:0036451) |
| 0.0 |
0.1 |
GO:0090220 |
meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 |
0.2 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.6 |
GO:0032291 |
central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 |
0.3 |
GO:0006048 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 |
0.5 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
1.0 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.2 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 |
0.2 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.1 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 |
1.1 |
GO:0031572 |
G2 DNA damage checkpoint(GO:0031572) |
| 0.0 |
0.2 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 |
0.9 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 |
0.1 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.0 |
0.3 |
GO:1903867 |
chorion development(GO:0060717) extraembryonic membrane development(GO:1903867) |
| 0.0 |
0.3 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.2 |
GO:0046686 |
response to cadmium ion(GO:0046686) |
| 0.0 |
0.7 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |
| 0.0 |
0.8 |
GO:0048741 |
skeletal muscle fiber development(GO:0048741) |
| 0.0 |
0.3 |
GO:0045589 |
regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 |
0.2 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.7 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.2 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
1.3 |
GO:0048663 |
neuron fate commitment(GO:0048663) |
| 0.0 |
0.2 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.1 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.0 |
0.2 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.5 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.4 |
GO:0006910 |
phagocytosis, recognition(GO:0006910) |
| 0.0 |
0.1 |
GO:0042832 |
response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.0 |
0.2 |
GO:0009437 |
carnitine metabolic process(GO:0009437) |
| 0.0 |
0.0 |
GO:0035166 |
post-embryonic hemopoiesis(GO:0035166) |
| 0.0 |
0.1 |
GO:1900225 |
NLRP3 inflammasome complex assembly(GO:0044546) regulation of NLRP3 inflammasome complex assembly(GO:1900225) |
| 0.0 |
0.1 |
GO:0044778 |
meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 |
0.3 |
GO:0021683 |
cerebellar granular layer morphogenesis(GO:0021683) |
| 0.0 |
0.2 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.2 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
0.1 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.0 |
0.1 |
GO:0086036 |
regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 |
0.1 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.4 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.3 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.0 |
0.1 |
GO:1902065 |
response to L-glutamate(GO:1902065) |
| 0.0 |
0.1 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.1 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 |
0.5 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.0 |
0.3 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.2 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.0 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 |
0.2 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.8 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.0 |
0.1 |
GO:1900037 |
regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 |
0.3 |
GO:0030033 |
microvillus assembly(GO:0030033) |
| 0.0 |
0.0 |
GO:0010881 |
regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.0 |
0.2 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 |
0.2 |
GO:0035999 |
tetrahydrofolate interconversion(GO:0035999) |
| 0.0 |
0.1 |
GO:0098751 |
bone cell development(GO:0098751) |
| 0.0 |
0.1 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.0 |
0.3 |
GO:0071353 |
response to interleukin-4(GO:0070670) cellular response to interleukin-4(GO:0071353) |
| 0.0 |
0.1 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 |
0.1 |
GO:0000271 |
polysaccharide biosynthetic process(GO:0000271) |
| 0.0 |
0.2 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 |
0.0 |
GO:0006094 |
gluconeogenesis(GO:0006094) |
| 0.0 |
0.1 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.0 |
0.0 |
GO:0097491 |
trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 |
0.0 |
GO:2000254 |
regulation of male germ cell proliferation(GO:2000254) |
| 0.0 |
0.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.4 |
GO:1902600 |
hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 |
0.2 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 |
0.1 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.0 |
0.1 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 |
0.1 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.0 |
0.1 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 |
0.1 |
GO:0023041 |
neuronal signal transduction(GO:0023041) |
| 0.0 |
0.1 |
GO:0044130 |
negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 |
0.1 |
GO:0051208 |
sequestering of calcium ion(GO:0051208) release of sequestered calcium ion into cytosol(GO:0051209) regulation of sequestering of calcium ion(GO:0051282) negative regulation of sequestering of calcium ion(GO:0051283) |
| 0.0 |
0.3 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.1 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.2 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.6 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.7 |
GO:0030879 |
mammary gland development(GO:0030879) |
| 0.0 |
0.0 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 |
0.3 |
GO:0009142 |
nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 |
0.3 |
GO:0055072 |
iron ion homeostasis(GO:0055072) |
| 0.0 |
0.1 |
GO:0006862 |
nucleotide transport(GO:0006862) |
| 0.0 |
0.1 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 |
0.5 |
GO:0000079 |
regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 |
0.8 |
GO:0007052 |
mitotic spindle organization(GO:0007052) |
| 0.0 |
0.1 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 |
0.2 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.0 |
0.1 |
GO:0008611 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 |
0.1 |
GO:0090201 |
negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 |
0.0 |
GO:0072539 |
T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) |
| 0.0 |
0.0 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.0 |
GO:0031590 |
wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 |
0.1 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.1 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.4 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.3 |
GO:0048286 |
lung alveolus development(GO:0048286) |
| 0.0 |
0.0 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.1 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 |
0.5 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
0.0 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |