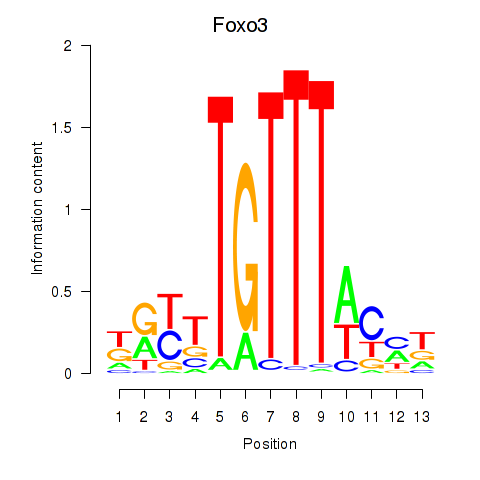
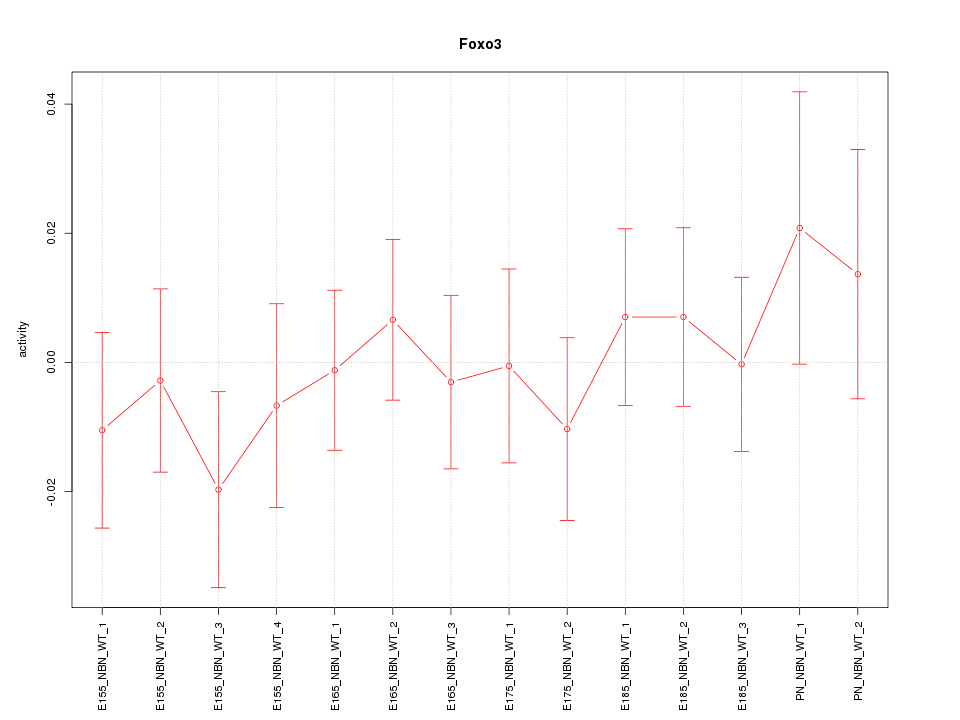
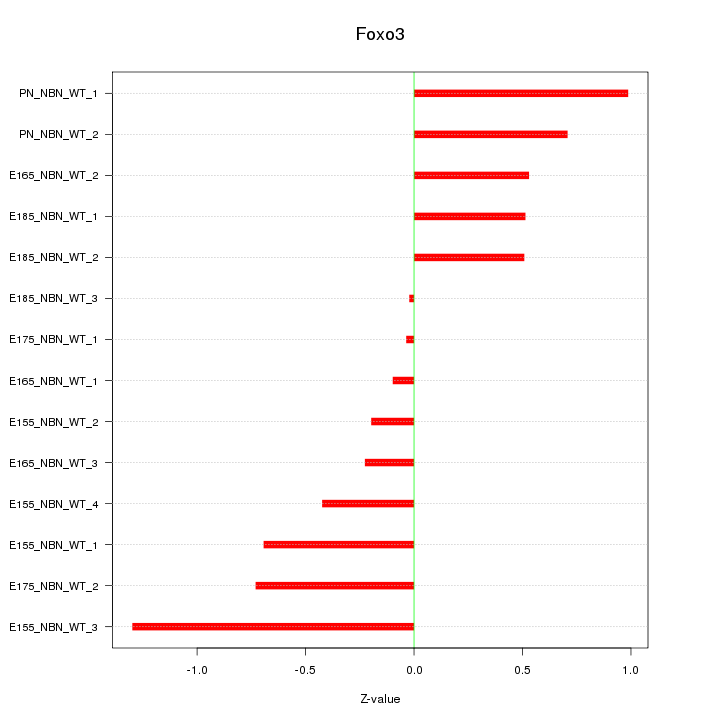
| 0.7 |
2.0 |
GO:1902524 |
negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.4 |
1.8 |
GO:0032901 |
positive regulation of neurotrophin production(GO:0032901) |
| 0.4 |
2.2 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.3 |
2.0 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.3 |
0.8 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.2 |
1.2 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.2 |
0.6 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.2 |
1.5 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.2 |
0.7 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 |
1.0 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
1.3 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
0.4 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.1 |
1.5 |
GO:0009109 |
coenzyme catabolic process(GO:0009109) |
| 0.1 |
0.4 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 |
0.3 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 |
1.5 |
GO:0010574 |
regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.1 |
0.3 |
GO:0006659 |
phosphatidylserine biosynthetic process(GO:0006659) cardiolipin biosynthetic process(GO:0032049) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 |
0.3 |
GO:0045908 |
negative regulation of vasodilation(GO:0045908) |
| 0.0 |
0.5 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.2 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
1.5 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.4 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 |
0.3 |
GO:2000807 |
regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 |
0.1 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 |
0.1 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 |
0.1 |
GO:0006083 |
acetate metabolic process(GO:0006083) |
| 0.0 |
0.2 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 |
1.0 |
GO:0048747 |
muscle fiber development(GO:0048747) |
| 0.0 |
0.1 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 |
0.0 |
GO:2000551 |
regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 |
0.5 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.0 |
0.1 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |