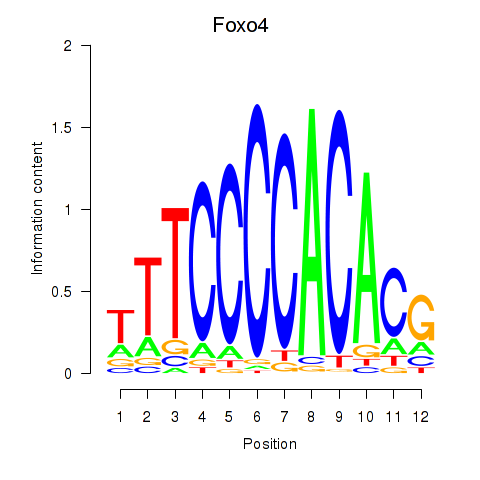
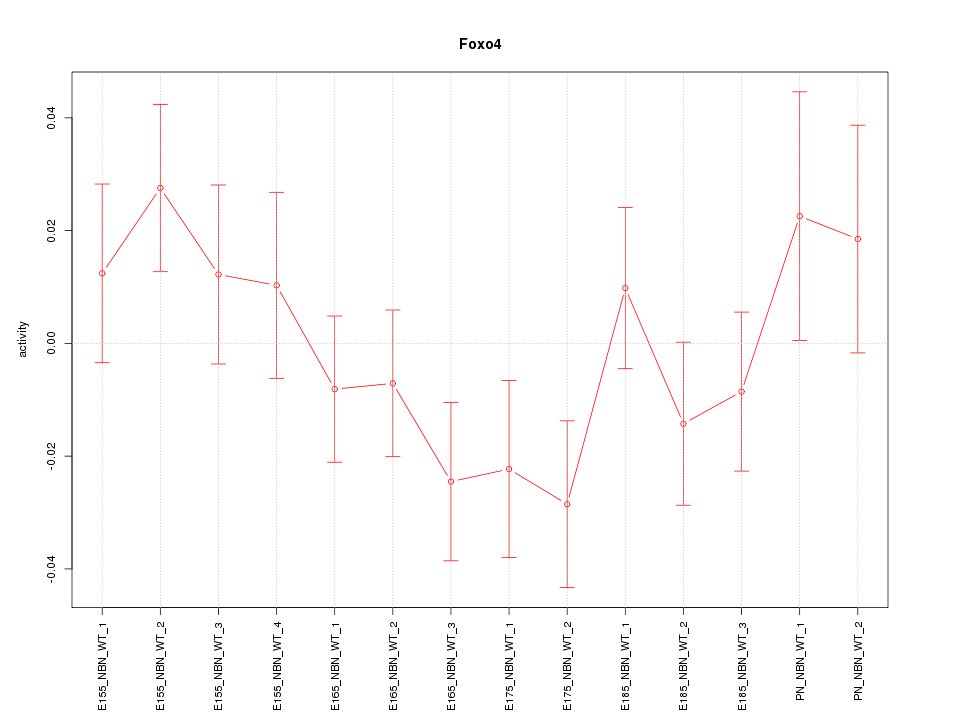
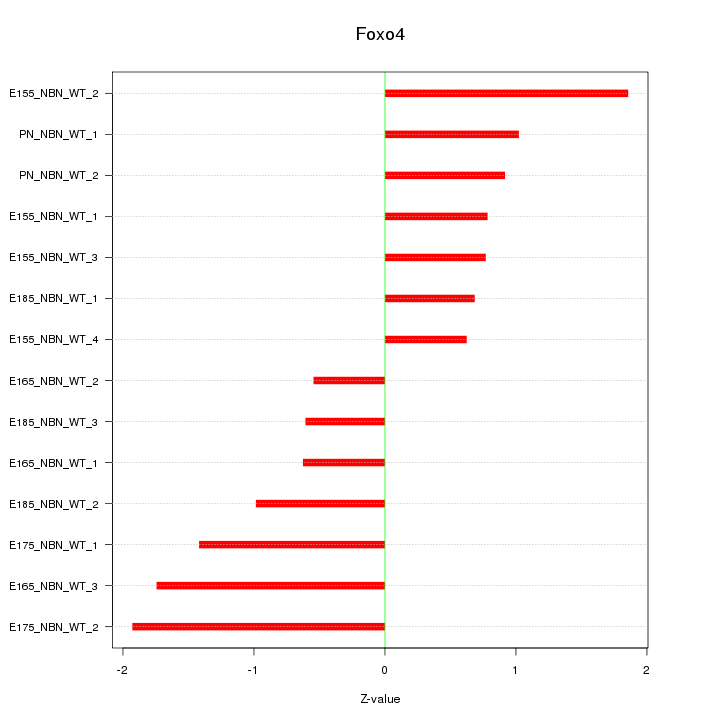
| 0.2 |
0.7 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 |
0.5 |
GO:0003104 |
positive regulation of glomerular filtration(GO:0003104) negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) phenotypic switching(GO:0036166) dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.1 |
1.6 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.3 |
GO:0045349 |
regulation of dendritic cell cytokine production(GO:0002730) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 |
1.2 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.3 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 |
0.3 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 |
1.4 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 |
0.2 |
GO:0032829 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) negative regulation of interleukin-2 biosynthetic process(GO:0045085) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 |
0.3 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
0.9 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 |
0.7 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 |
0.2 |
GO:0002314 |
germinal center B cell differentiation(GO:0002314) |
| 0.1 |
0.4 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.0 |
1.8 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
0.2 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 |
0.3 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.0 |
0.2 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 |
0.2 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.3 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.5 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.0 |
0.6 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 |
0.2 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 |
0.2 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |