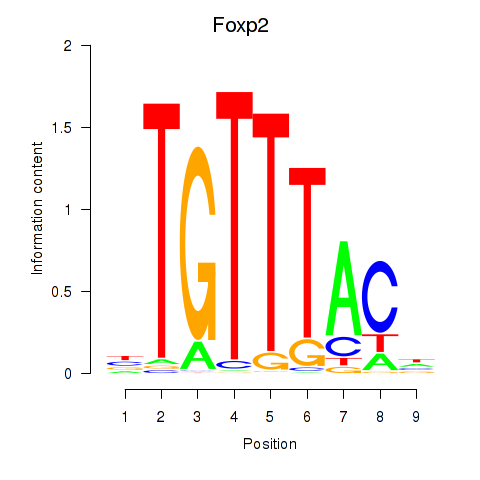
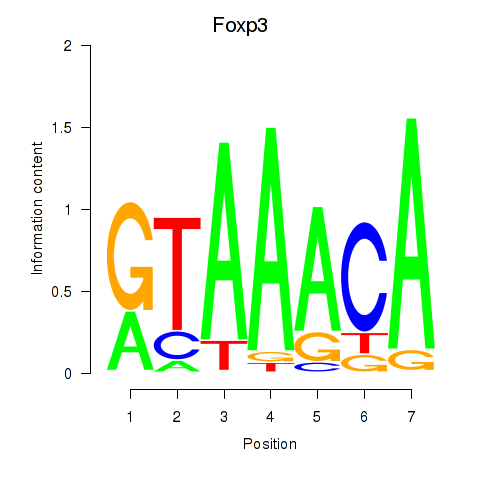
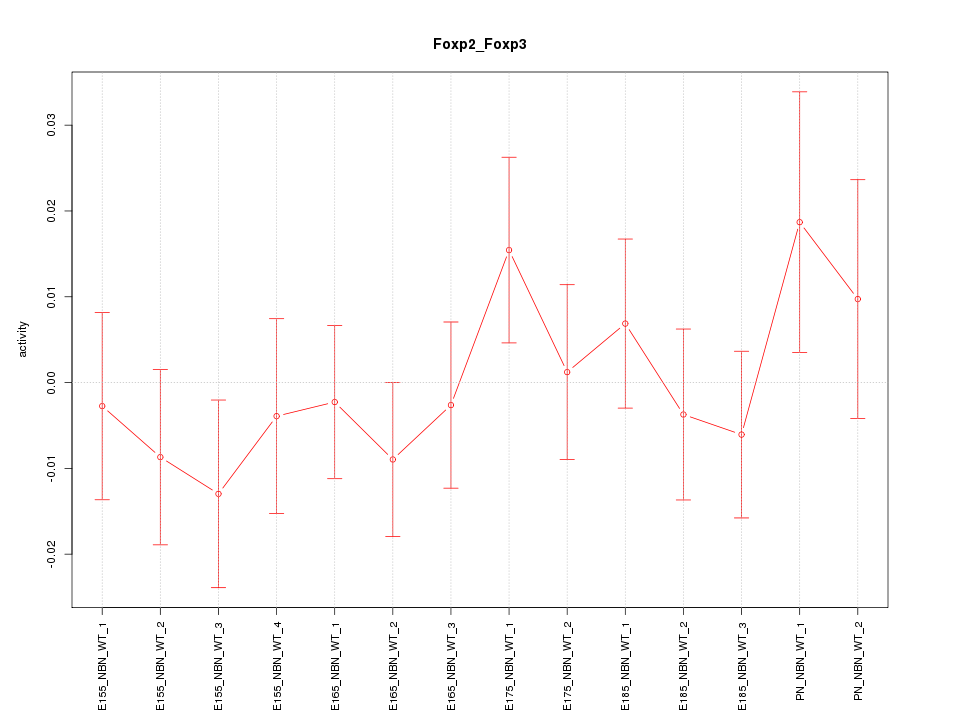
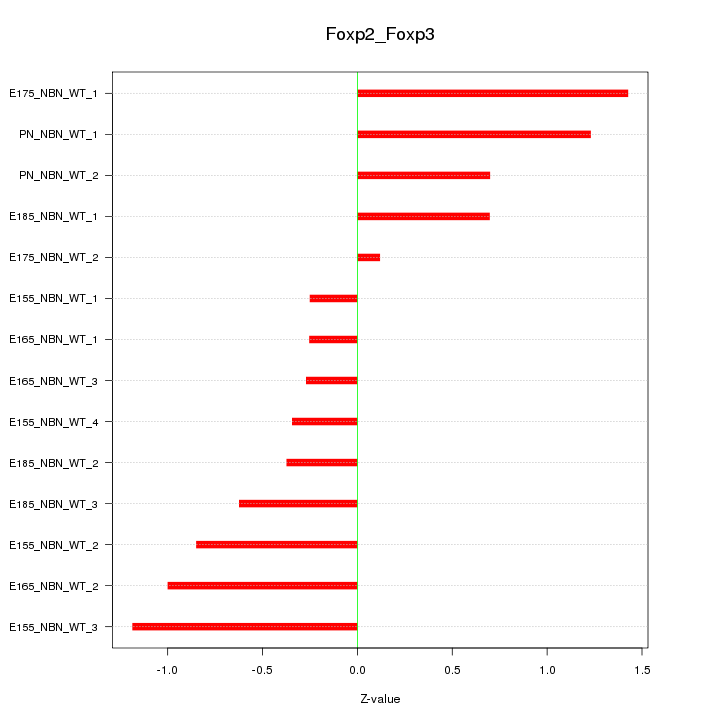
| 0.7 |
8.2 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.7 |
2.2 |
GO:1901254 |
positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.5 |
1.0 |
GO:2000078 |
positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.4 |
1.3 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.4 |
1.3 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.4 |
1.2 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.4 |
2.5 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.3 |
1.4 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.3 |
1.0 |
GO:1902524 |
negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.3 |
3.1 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.3 |
0.6 |
GO:0072076 |
nephrogenic mesenchyme development(GO:0072076) |
| 0.3 |
3.1 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.3 |
0.8 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 |
2.2 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.3 |
0.8 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 |
4.4 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 |
1.2 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.2 |
2.1 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.2 |
0.6 |
GO:0086069 |
desmosome assembly(GO:0002159) bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.2 |
0.6 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.2 |
1.0 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 |
0.9 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.2 |
0.7 |
GO:0055099 |
regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 0.2 |
0.7 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.2 |
0.7 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 |
0.7 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 |
2.7 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.2 |
0.2 |
GO:0000821 |
regulation of arginine metabolic process(GO:0000821) |
| 0.2 |
2.4 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.2 |
0.3 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.2 |
2.6 |
GO:0097154 |
GABAergic neuron differentiation(GO:0097154) |
| 0.1 |
0.9 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 |
0.6 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 |
0.8 |
GO:2000973 |
regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 |
2.5 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 |
1.4 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.1 |
1.2 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.1 |
0.7 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.1 |
0.8 |
GO:0050861 |
regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.8 |
GO:0021984 |
adenohypophysis development(GO:0021984) |
| 0.1 |
2.8 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 |
1.5 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 |
0.4 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 |
0.4 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) Notch signaling involved in heart development(GO:0061314) |
| 0.1 |
1.0 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
1.0 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.1 |
0.3 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
0.4 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.1 |
0.3 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 |
0.5 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 |
0.7 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.1 |
0.7 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 |
0.9 |
GO:0030538 |
embryonic genitalia morphogenesis(GO:0030538) embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 |
0.2 |
GO:2000668 |
dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.1 |
0.7 |
GO:0032264 |
IMP salvage(GO:0032264) |
| 0.1 |
0.3 |
GO:1903054 |
negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 |
0.5 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 |
0.5 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 |
0.8 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.3 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 |
0.6 |
GO:0051461 |
protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 |
1.0 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.1 |
0.3 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 |
0.3 |
GO:2000675 |
negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 |
1.1 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 |
0.3 |
GO:0071930 |
negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 |
0.5 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.1 |
0.5 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.1 |
0.3 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 |
0.4 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
0.5 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.1 |
0.1 |
GO:1905005 |
regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.1 |
0.2 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 |
0.2 |
GO:0071104 |
response to interleukin-9(GO:0071104) |
| 0.1 |
0.3 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
1.9 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
0.6 |
GO:0010572 |
positive regulation of platelet activation(GO:0010572) |
| 0.1 |
1.0 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.1 |
0.3 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.1 |
0.5 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
0.3 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.1 |
1.5 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 |
0.6 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 |
1.1 |
GO:0010574 |
regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.1 |
0.4 |
GO:0055003 |
cardiac myofibril assembly(GO:0055003) |
| 0.1 |
0.5 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.3 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.0 |
0.6 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.2 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.0 |
0.3 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.2 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.1 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 |
0.2 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.0 |
0.2 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 |
0.6 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.1 |
GO:0060913 |
cardiac cell fate determination(GO:0060913) regulation of cardiac cell fate specification(GO:2000043) |
| 0.0 |
0.9 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.3 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 |
0.3 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.4 |
GO:0002643 |
regulation of tolerance induction(GO:0002643) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.7 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.5 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.1 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 |
0.7 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 |
0.2 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.2 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) |
| 0.0 |
0.1 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.0 |
1.4 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.2 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.3 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.1 |
GO:0072488 |
nitrogen utilization(GO:0019740) ammonium transmembrane transport(GO:0072488) |
| 0.0 |
0.1 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.0 |
0.2 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.0 |
0.5 |
GO:0010592 |
positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 |
0.1 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.0 |
0.3 |
GO:0043970 |
histone H3-K9 acetylation(GO:0043970) |
| 0.0 |
0.1 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 |
0.1 |
GO:0071033 |
nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 |
0.1 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.0 |
0.2 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.0 |
0.2 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 |
0.2 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.0 |
1.6 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.0 |
0.5 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.3 |
GO:0071578 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.1 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.0 |
0.8 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.3 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.3 |
GO:0071168 |
protein localization to chromatin(GO:0071168) |
| 0.0 |
0.6 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.2 |
GO:0002347 |
response to tumor cell(GO:0002347) |
| 0.0 |
0.4 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.2 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.0 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 |
0.3 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.1 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 |
0.3 |
GO:0021684 |
cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.1 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.0 |
0.2 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 |
1.2 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.1 |
GO:2000857 |
positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 |
0.1 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.0 |
0.1 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 |
0.8 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.1 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 |
0.0 |
GO:0043987 |
histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 |
0.6 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.1 |
GO:0002544 |
chronic inflammatory response(GO:0002544) |
| 0.0 |
0.4 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.0 |
0.3 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
0.3 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.0 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 |
0.1 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 |
0.8 |
GO:0030071 |
regulation of mitotic metaphase/anaphase transition(GO:0030071) regulation of metaphase/anaphase transition of cell cycle(GO:1902099) |
| 0.0 |
0.3 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.0 |
0.1 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.0 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 |
0.1 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.3 |
GO:1904355 |
positive regulation of telomere capping(GO:1904355) |
| 0.0 |
0.2 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.0 |
0.2 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.4 |
GO:0010972 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 |
0.3 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.3 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.1 |
GO:2000598 |
regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 |
0.1 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.4 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.1 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 |
0.1 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.1 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 |
0.1 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 |
0.1 |
GO:0002693 |
positive regulation of cellular extravasation(GO:0002693) |
| 0.0 |
0.1 |
GO:0097460 |
ferrous iron import(GO:0070627) ferrous iron import into cell(GO:0097460) |
| 0.0 |
0.1 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.0 |
0.3 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.5 |
GO:0030500 |
regulation of bone mineralization(GO:0030500) |
| 0.0 |
0.1 |
GO:0007618 |
mating behavior(GO:0007617) mating(GO:0007618) multi-organism reproductive behavior(GO:0044705) |