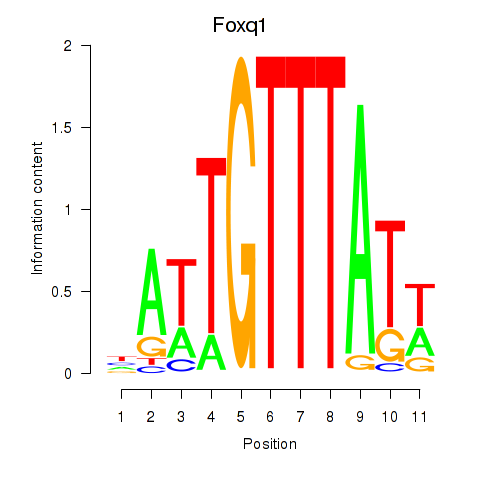
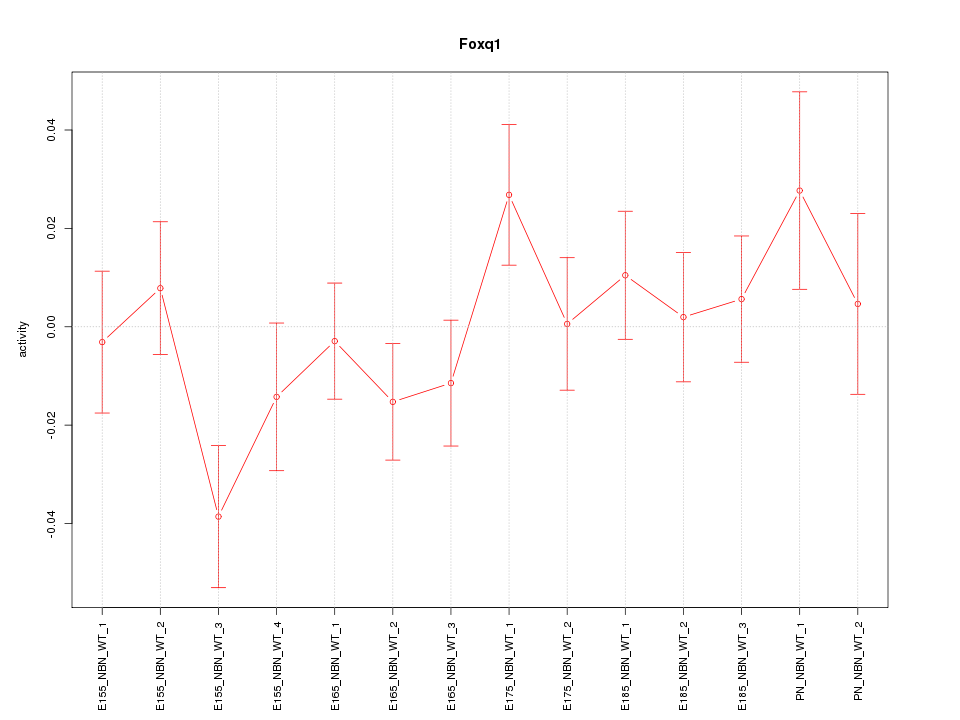
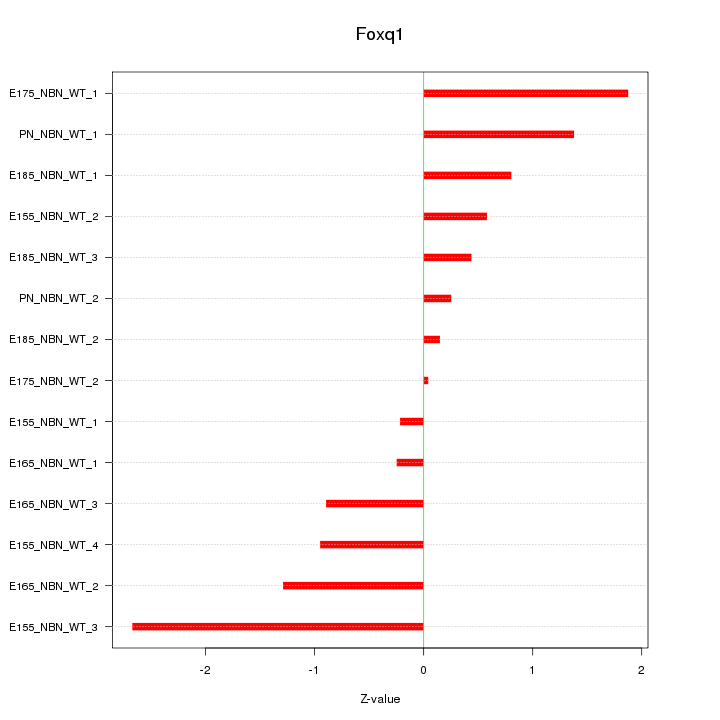
| 1.7 |
5.2 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.8 |
2.5 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.7 |
2.0 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 0.3 |
0.9 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) |
| 0.3 |
0.9 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.2 |
1.7 |
GO:0032264 |
IMP salvage(GO:0032264) |
| 0.2 |
0.6 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.2 |
0.6 |
GO:0072076 |
nephrogenic mesenchyme development(GO:0072076) |
| 0.2 |
1.8 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.2 |
0.5 |
GO:0046544 |
regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) development of secondary male sexual characteristics(GO:0046544) |
| 0.1 |
0.8 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.1 |
0.4 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 |
0.6 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.1 |
0.8 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 |
3.3 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.2 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 |
0.2 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.1 |
0.2 |
GO:0097501 |
stress response to metal ion(GO:0097501) |
| 0.1 |
0.4 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.4 |
GO:0035234 |
ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 |
0.8 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.6 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.6 |
GO:0002643 |
regulation of tolerance induction(GO:0002643) |
| 0.1 |
0.1 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 |
0.5 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 |
1.0 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.1 |
0.5 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 |
0.1 |
GO:0072095 |
regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 |
0.4 |
GO:2000301 |
myeloid progenitor cell differentiation(GO:0002318) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 |
0.2 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.0 |
4.8 |
GO:0072332 |
intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
0.3 |
GO:0032196 |
transposition(GO:0032196) |
| 0.0 |
1.1 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 |
1.3 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
0.2 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 |
0.3 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.1 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.0 |
0.4 |
GO:0006825 |
copper ion transport(GO:0006825) response to copper ion(GO:0046688) |
| 0.0 |
0.7 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
1.0 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.0 |
0.2 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.2 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.3 |
GO:0060484 |
lung-associated mesenchyme development(GO:0060484) |
| 0.0 |
0.1 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.0 |
0.2 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.0 |
0.1 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.1 |
GO:0046210 |
nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.0 |
0.2 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.3 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.3 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.9 |
GO:0055072 |
iron ion homeostasis(GO:0055072) |
| 0.0 |
1.0 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:0032790 |
ribosome disassembly(GO:0032790) |