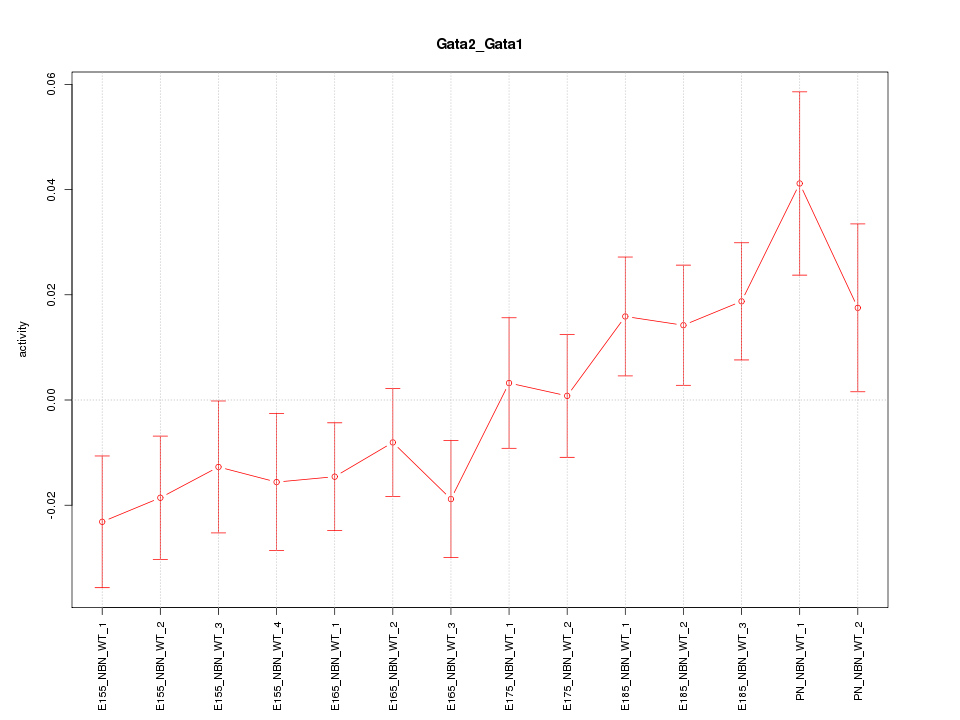
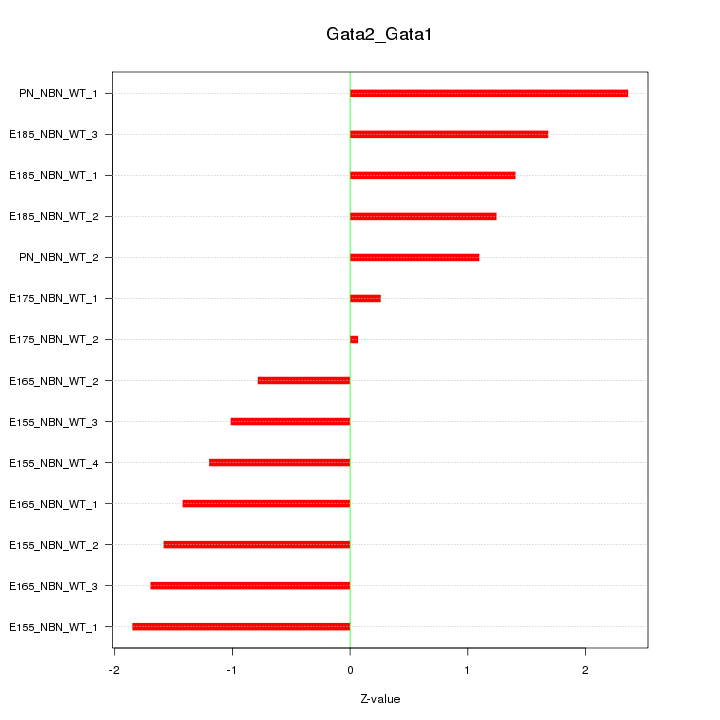
Motif ID: Gata2_Gata1
Z-value: 1.392
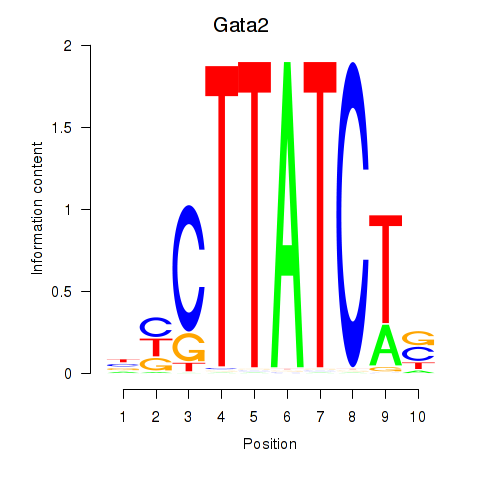
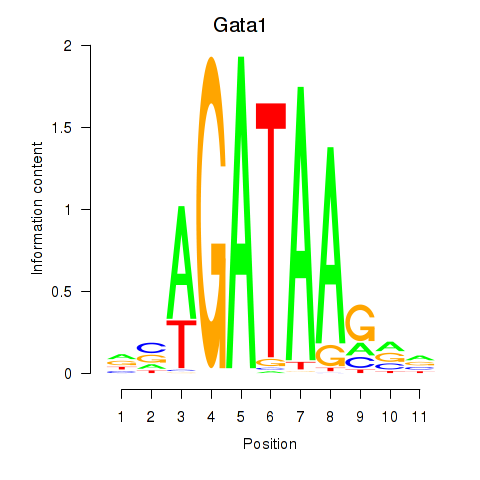
Transcription factors associated with Gata2_Gata1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Gata1 | ENSMUSG00000031162.8 | Gata1 |
| Gata2 | ENSMUSG00000015053.8 | Gata2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata2 | mm10_v2_chr6_+_88198656_88198675 | 0.80 | 5.6e-04 | Click! |
| Gata1 | mm10_v2_chrX_-_7967817_7967910 | 0.33 | 2.5e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 1.0 | 7.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.9 | 5.1 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.7 | 3.7 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.6 | 6.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.6 | 6.1 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.6 | 1.7 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.5 | 5.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.5 | 3.2 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.5 | 1.5 | GO:0015920 | regulation of phosphatidylcholine catabolic process(GO:0010899) lipopolysaccharide transport(GO:0015920) |
| 0.4 | 1.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.4 | 1.2 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.4 | 2.6 | GO:1901162 | tryptophan metabolic process(GO:0006568) indolalkylamine metabolic process(GO:0006586) serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.4 | 2.5 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 1.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.3 | 1.9 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.3 | 1.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 0.9 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.3 | 1.1 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.3 | 1.3 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.2 | 1.5 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 2.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 2.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 2.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 0.5 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.2 | 1.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 0.8 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.2 | 0.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 1.5 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 0.4 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 | 1.9 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 1.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.8 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 2.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.6 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.5 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 2.1 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 1.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.3 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.4 | GO:0097168 | condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 | 0.2 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 2.9 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 1.0 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 1.5 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 | 1.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 2.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 | 5.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 4.7 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 1.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.5 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 1.3 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.3 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 1.1 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.3 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 1.6 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 1.6 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 1.2 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 1.5 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.3 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.7 | 3.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 1.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.3 | 0.8 | GO:0043512 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.3 | 4.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.7 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 1.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 2.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 1.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.0 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.4 | GO:0071438 | NADPH oxidase complex(GO:0043020) invadopodium membrane(GO:0071438) |
| 0.1 | 1.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.6 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 1.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 3.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.3 | GO:0000793 | condensed chromosome(GO:0000793) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 1.1 | 5.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.9 | 2.6 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.6 | 1.9 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.6 | 1.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.5 | 1.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.4 | 1.3 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.4 | 2.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 2.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 6.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 3.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.3 | 1.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) arginine binding(GO:0034618) |
| 0.3 | 3.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 1.2 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.3 | 1.5 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.3 | 1.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.2 | 5.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 6.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.7 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.2 | 1.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled adenosine receptor activity(GO:0001609) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 2.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 1.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 5.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.4 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.1 | 0.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.8 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 1.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.6 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 2.1 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 0.2 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 0.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 3.8 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 1.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.5 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 2.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 2.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 4.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 4.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |