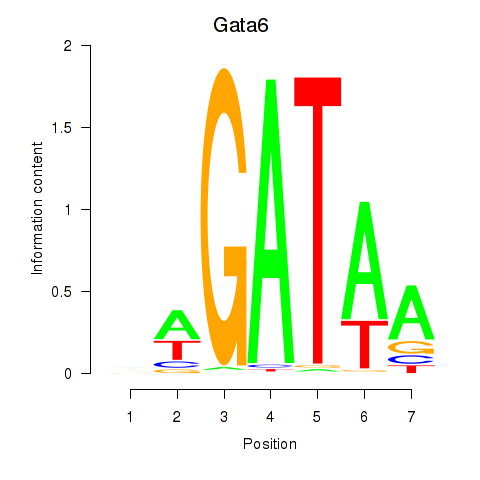
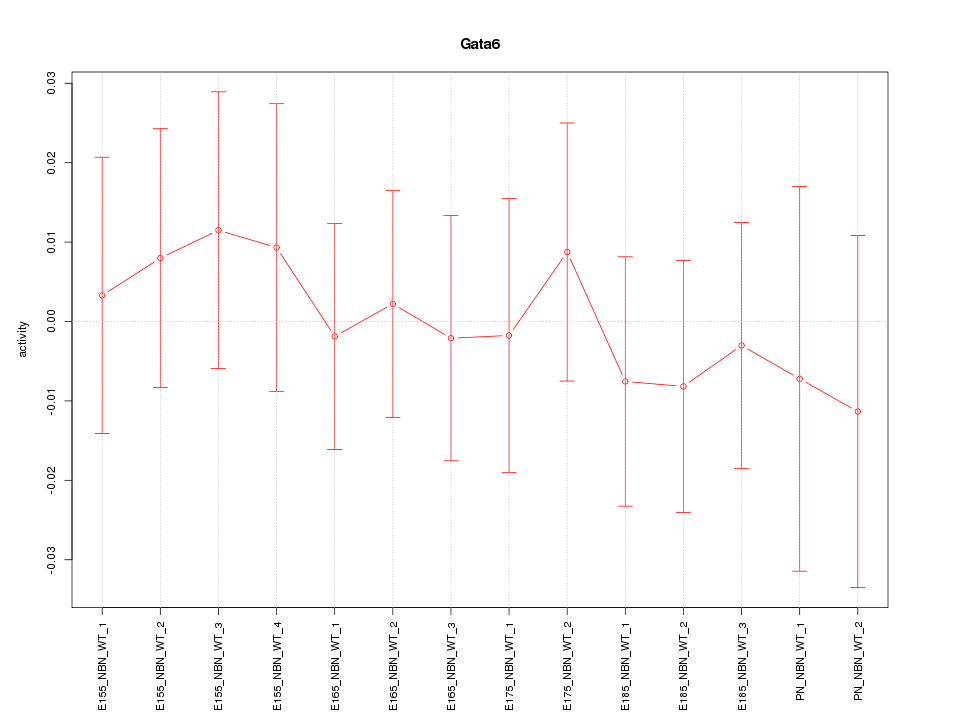
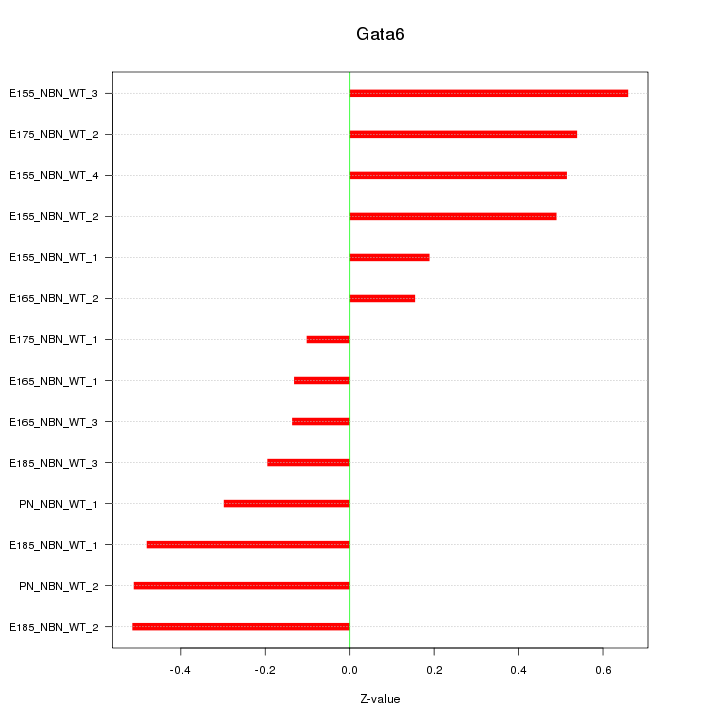
| 0.2 |
0.7 |
GO:0035878 |
nail development(GO:0035878) |
| 0.2 |
0.9 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.2 |
0.9 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 |
0.6 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.1 |
0.3 |
GO:0000019 |
regulation of mitotic recombination(GO:0000019) |
| 0.1 |
0.5 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.7 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.4 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 |
1.2 |
GO:0021692 |
cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 |
1.5 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.2 |
GO:0060414 |
aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 |
0.1 |
GO:0000350 |
generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 |
0.6 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.1 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 |
1.1 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.1 |
GO:0032380 |
regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 |
0.1 |
GO:0070072 |
vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 |
0.2 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.1 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.0 |
0.4 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.1 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 |
0.1 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.0 |
0.1 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 |
0.1 |
GO:0034242 |
negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 |
0.2 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.3 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.0 |
0.3 |
GO:0019432 |
triglyceride biosynthetic process(GO:0019432) |