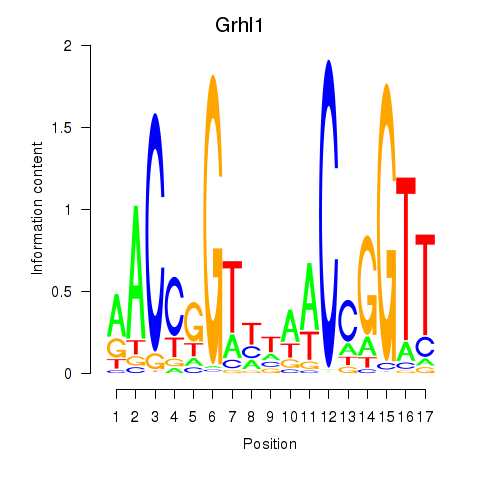
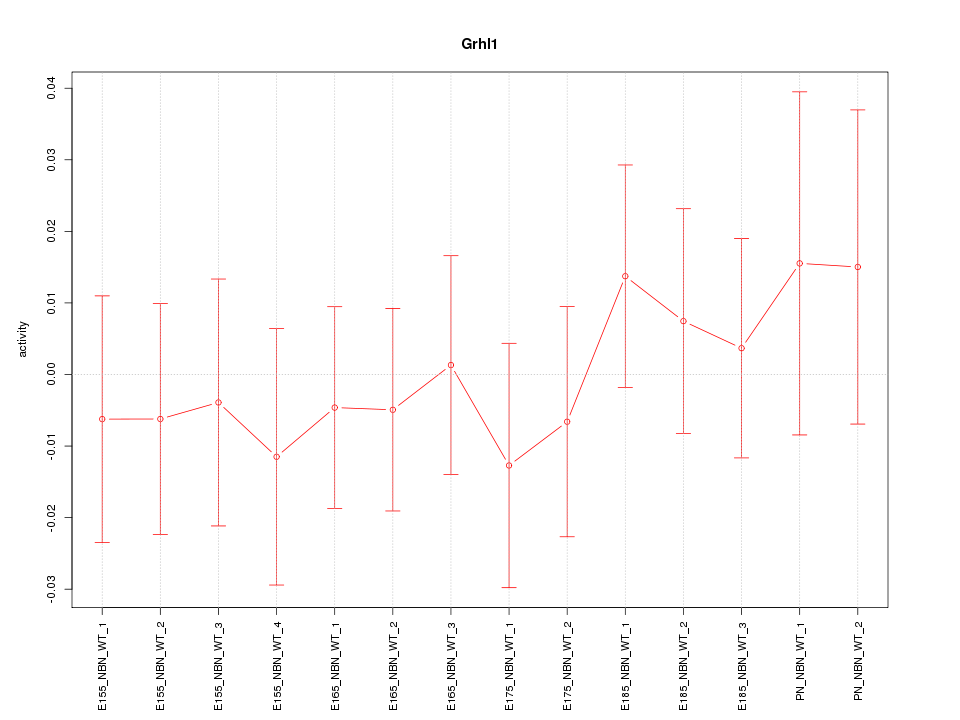
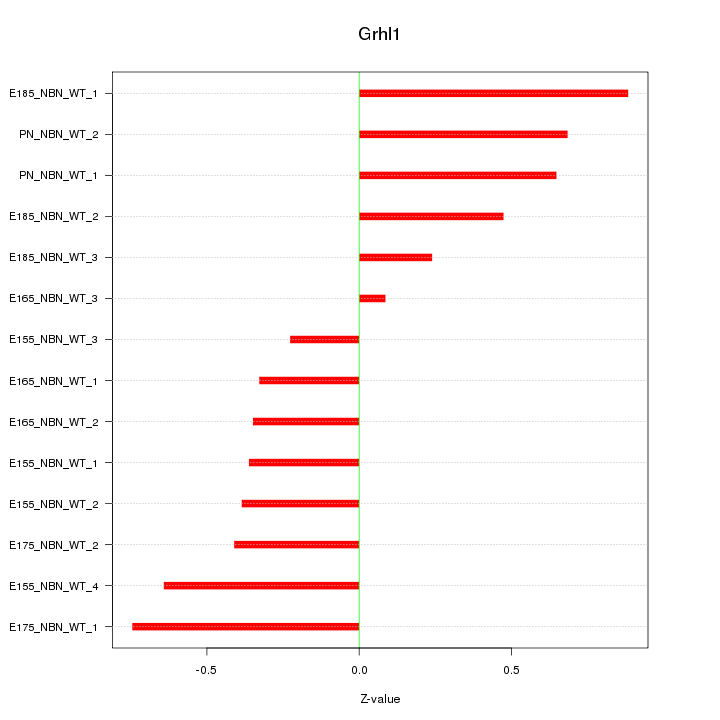
| 0.1 |
0.6 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 |
0.5 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 |
0.6 |
GO:0047484 |
regulation of response to osmotic stress(GO:0047484) |
| 0.1 |
0.7 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.9 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
0.3 |
GO:0042985 |
negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 |
0.2 |
GO:0032782 |
bile acid secretion(GO:0032782) positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 |
0.4 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 |
0.2 |
GO:0071340 |
skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 |
0.1 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.1 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.1 |
GO:1901727 |
positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 |
0.1 |
GO:1900242 |
regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 |
0.2 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.1 |
GO:0097341 |
inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 |
0.1 |
GO:1902866 |
regulation of retina development in camera-type eye(GO:1902866) |