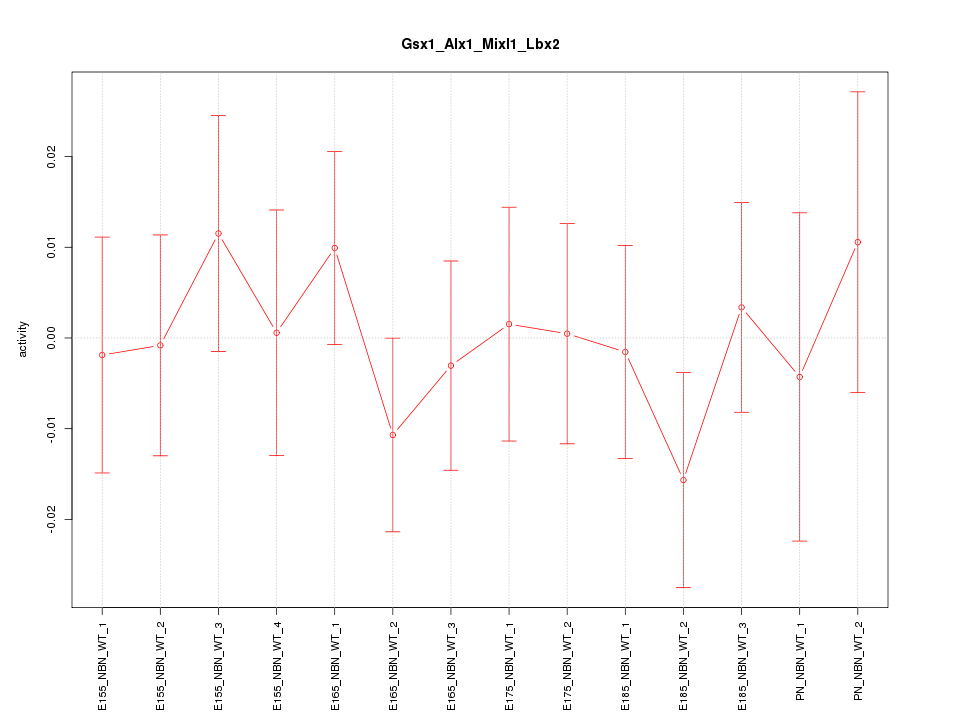
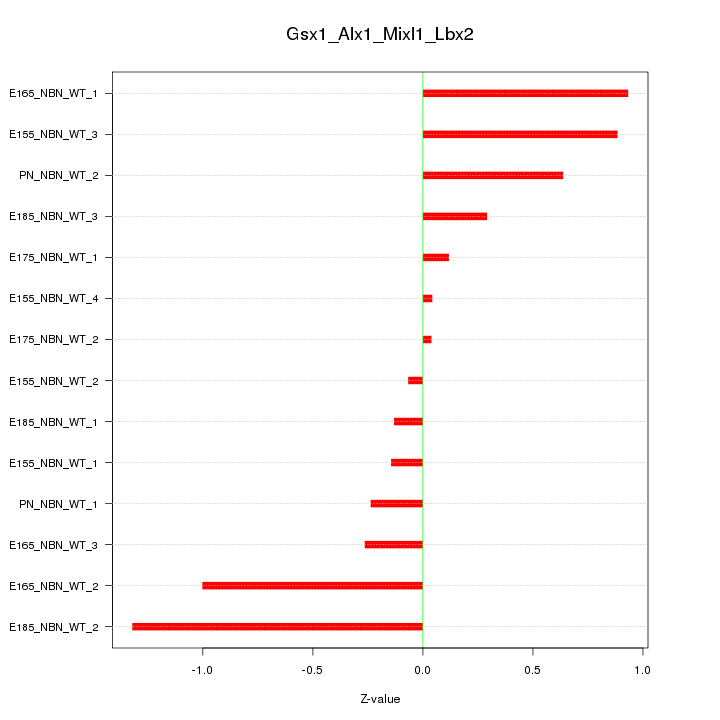
| 0.3 |
0.8 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.2 |
0.5 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.2 |
0.6 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 |
0.9 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 |
0.4 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 |
0.3 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.1 |
0.9 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 |
0.4 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 |
0.5 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
0.2 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 |
0.4 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.1 |
0.3 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.1 |
0.2 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
0.5 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 |
0.2 |
GO:1902605 |
heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.1 |
0.2 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 |
0.3 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 |
0.2 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 |
0.1 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 |
0.9 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.3 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.0 |
0.0 |
GO:0003162 |
atrioventricular node development(GO:0003162) |
| 0.0 |
0.3 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.1 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 |
0.0 |
GO:1903546 |
protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 |
0.2 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 |
0.2 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 |
0.2 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.0 |
0.1 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 |
0.5 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.4 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 |
0.6 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.6 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.1 |
GO:1904956 |
regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 |
0.1 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) |
| 0.0 |
0.3 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.1 |
GO:0090298 |
negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.0 |
0.1 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 |
0.2 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.0 |
0.2 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.0 |
0.2 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.0 |
0.3 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.4 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.2 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 |
0.1 |
GO:1904742 |
protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.0 |
0.6 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.0 |
0.1 |
GO:0008627 |
intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 |
0.2 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 |
0.1 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 |
0.1 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 |
0.1 |
GO:0051798 |
positive regulation of hair follicle development(GO:0051798) |
| 0.0 |
0.4 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.4 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.2 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
0.1 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.6 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.1 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 |
0.1 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 |
0.4 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.0 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.0 |
0.1 |
GO:2000324 |
positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 |
0.1 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.0 |
0.3 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.0 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.0 |
0.0 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.0 |
0.0 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 |
0.5 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.1 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.3 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
0.2 |
GO:0010715 |
regulation of extracellular matrix disassembly(GO:0010715) |
| 0.0 |
0.2 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.0 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 |
0.0 |
GO:0016344 |
meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 |
0.0 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.4 |
GO:2000249 |
regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 |
0.1 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.0 |
GO:0045852 |
pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 |
0.1 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.3 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.0 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.0 |
0.0 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.0 |
0.1 |
GO:0021775 |
smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 |
0.1 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 |
0.0 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |