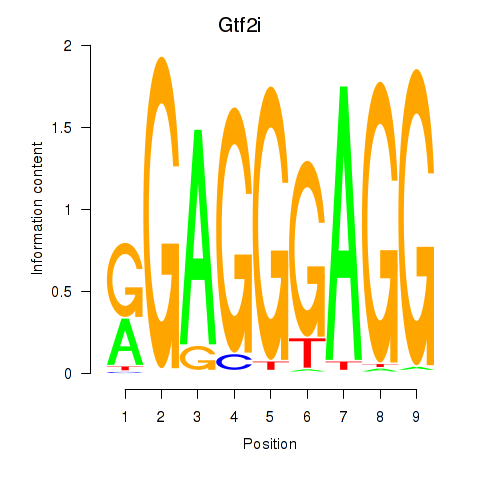
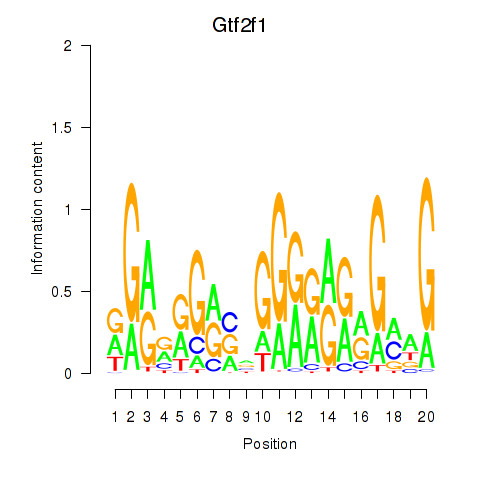
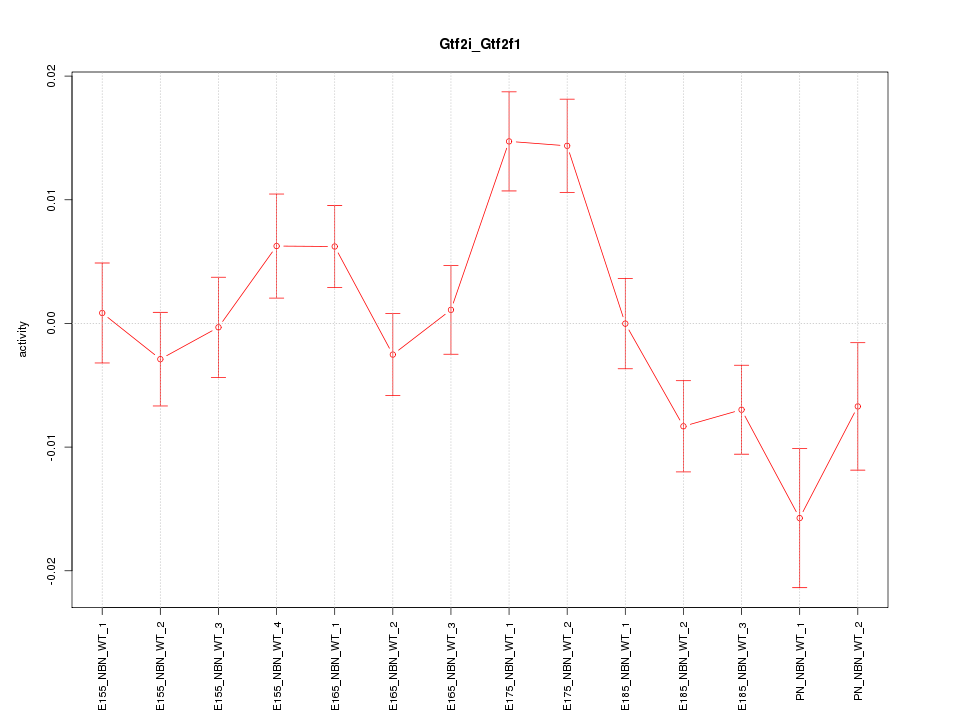
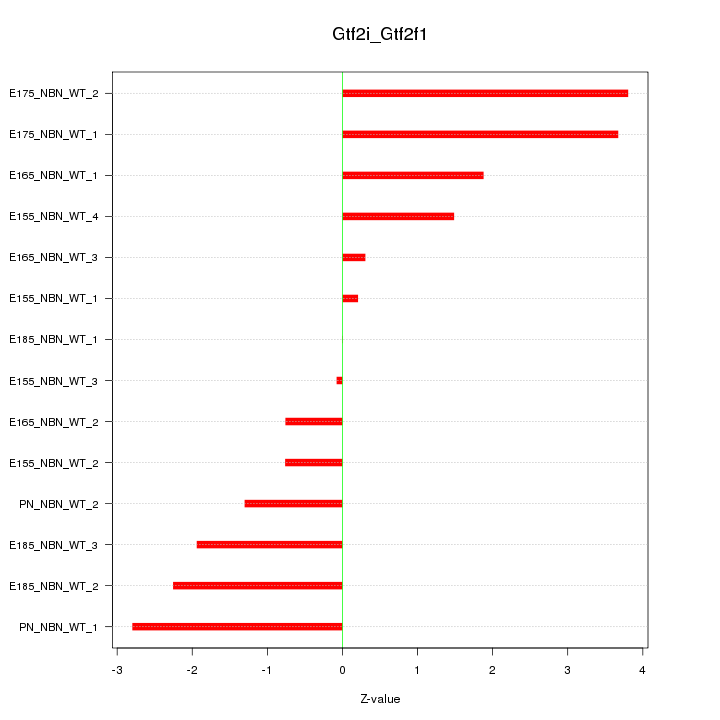
| 1.9 |
5.7 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.5 |
4.4 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 1.5 |
4.4 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 1.4 |
6.8 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 1.3 |
4.0 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 1.2 |
3.6 |
GO:0046959 |
habituation(GO:0046959) |
| 1.2 |
7.0 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 1.1 |
3.2 |
GO:0060023 |
soft palate development(GO:0060023) |
| 1.0 |
4.0 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.9 |
2.7 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.9 |
1.8 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.8 |
2.5 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.8 |
6.5 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.8 |
7.7 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.7 |
2.1 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.6 |
1.9 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.6 |
3.2 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.6 |
1.9 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.6 |
1.9 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.6 |
1.7 |
GO:0036015 |
response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.6 |
2.2 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.5 |
1.6 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.5 |
7.8 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.5 |
3.1 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.5 |
2.0 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.5 |
1.5 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.5 |
1.5 |
GO:2000813 |
actin filament uncapping(GO:0051695) negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.5 |
2.5 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.5 |
2.0 |
GO:0051305 |
chromosome movement towards spindle pole(GO:0051305) |
| 0.5 |
1.5 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.5 |
1.8 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.5 |
1.4 |
GO:0036353 |
histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.4 |
3.5 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.4 |
4.3 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.4 |
1.2 |
GO:0009644 |
response to high light intensity(GO:0009644) |
| 0.4 |
1.2 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.4 |
1.2 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.4 |
1.2 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.4 |
1.2 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.4 |
1.5 |
GO:1903966 |
monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.4 |
1.9 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.4 |
1.1 |
GO:0061535 |
amygdala development(GO:0021764) glutamate secretion, neurotransmission(GO:0061535) |
| 0.4 |
1.8 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.4 |
1.1 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.4 |
1.1 |
GO:1904049 |
regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.4 |
2.1 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.4 |
0.4 |
GO:2000286 |
receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.4 |
1.4 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.3 |
0.7 |
GO:0009957 |
epidermal cell fate specification(GO:0009957) |
| 0.3 |
2.8 |
GO:0097118 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.3 |
2.4 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 |
1.0 |
GO:0060915 |
fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) positive regulation of fat cell proliferation(GO:0070346) |
| 0.3 |
0.7 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 0.3 |
1.6 |
GO:0010616 |
negative regulation of cardiac muscle adaptation(GO:0010616) |
| 0.3 |
1.3 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.3 |
0.9 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 |
0.9 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.3 |
0.9 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.3 |
1.9 |
GO:0061669 |
spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.3 |
1.2 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 |
2.5 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.3 |
5.1 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.3 |
1.2 |
GO:0033024 |
mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.3 |
1.7 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) protein localization to ciliary transition zone(GO:1904491) |
| 0.3 |
0.6 |
GO:0070212 |
protein poly-ADP-ribosylation(GO:0070212) |
| 0.3 |
0.8 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.3 |
0.8 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.3 |
2.2 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.3 |
1.7 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 |
1.4 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.3 |
0.8 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 |
1.6 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 |
1.1 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.3 |
0.8 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.3 |
1.3 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.3 |
0.3 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 |
1.3 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 |
1.0 |
GO:2000546 |
positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.2 |
2.2 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 |
3.4 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 |
0.2 |
GO:0033292 |
T-tubule organization(GO:0033292) |
| 0.2 |
0.7 |
GO:0048505 |
regulation of timing of cell differentiation(GO:0048505) |
| 0.2 |
3.6 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.2 |
0.2 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.2 |
0.2 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.2 |
0.2 |
GO:0070671 |
response to interleukin-12(GO:0070671) |
| 0.2 |
0.9 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 |
0.7 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.2 |
1.1 |
GO:0051012 |
microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.2 |
5.0 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.2 |
1.1 |
GO:0034436 |
glycoprotein transport(GO:0034436) |
| 0.2 |
0.7 |
GO:0033364 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mast cell secretory granule organization(GO:0033364) |
| 0.2 |
1.3 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.2 |
1.3 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 |
0.6 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 |
1.1 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.2 |
0.2 |
GO:2001015 |
negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.2 |
0.8 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.2 |
2.9 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.2 |
1.0 |
GO:0000972 |
transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.2 |
1.8 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.2 |
1.0 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 |
1.0 |
GO:0007403 |
glial cell fate determination(GO:0007403) |
| 0.2 |
1.0 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 |
1.6 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.2 |
0.6 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.2 |
0.2 |
GO:0042713 |
sperm ejaculation(GO:0042713) |
| 0.2 |
0.4 |
GO:0070885 |
negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.2 |
0.4 |
GO:0019042 |
viral latency(GO:0019042) |
| 0.2 |
0.8 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.2 |
0.2 |
GO:2000807 |
regulation of synaptic vesicle clustering(GO:2000807) |
| 0.2 |
0.6 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.2 |
1.3 |
GO:0014824 |
artery smooth muscle contraction(GO:0014824) |
| 0.2 |
1.3 |
GO:0060087 |
relaxation of vascular smooth muscle(GO:0060087) |
| 0.2 |
0.9 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 |
1.1 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.2 |
1.6 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.2 |
0.4 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.2 |
0.7 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 |
0.5 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.2 |
0.4 |
GO:1901492 |
positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 |
1.1 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 |
0.4 |
GO:0010752 |
regulation of cGMP-mediated signaling(GO:0010752) |
| 0.2 |
0.4 |
GO:1904529 |
regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.2 |
0.4 |
GO:0033563 |
dorsal/ventral axon guidance(GO:0033563) |
| 0.2 |
1.1 |
GO:0090188 |
negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.2 |
0.7 |
GO:1900623 |
regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 |
0.9 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 0.2 |
1.6 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.2 |
0.2 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.2 |
0.9 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.2 |
0.7 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.2 |
4.3 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.2 |
1.0 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.2 |
0.5 |
GO:2001206 |
positive regulation of osteoclast development(GO:2001206) |
| 0.2 |
2.2 |
GO:0015732 |
prostaglandin transport(GO:0015732) |
| 0.2 |
1.2 |
GO:0051775 |
response to redox state(GO:0051775) |
| 0.2 |
1.8 |
GO:0090331 |
negative regulation of platelet aggregation(GO:0090331) |
| 0.2 |
0.3 |
GO:0014735 |
regulation of muscle atrophy(GO:0014735) |
| 0.2 |
1.0 |
GO:0060576 |
intestinal epithelial cell development(GO:0060576) |
| 0.2 |
0.5 |
GO:0019659 |
glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.2 |
0.8 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.2 |
0.5 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) |
| 0.2 |
2.7 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.2 |
0.5 |
GO:0035973 |
aggrephagy(GO:0035973) |
| 0.2 |
2.1 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.2 |
0.3 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.2 |
0.8 |
GO:0010807 |
regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 |
0.5 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 |
1.3 |
GO:1902669 |
positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.2 |
0.5 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 |
0.8 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.2 |
0.3 |
GO:0097051 |
establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 |
0.4 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.1 |
0.7 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 |
0.9 |
GO:0060510 |
Type II pneumocyte differentiation(GO:0060510) |
| 0.1 |
1.0 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
1.5 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 |
0.6 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 |
0.6 |
GO:0006529 |
asparagine biosynthetic process(GO:0006529) |
| 0.1 |
1.9 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.4 |
GO:0086103 |
G-protein coupled receptor signaling pathway involved in heart process(GO:0086103) |
| 0.1 |
0.6 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.1 |
0.4 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 |
1.1 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.1 |
1.4 |
GO:0021957 |
corticospinal tract morphogenesis(GO:0021957) |
| 0.1 |
0.4 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.1 |
2.4 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.1 |
0.4 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 |
0.1 |
GO:2000170 |
positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.1 |
1.4 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 |
0.8 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
1.1 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 |
1.2 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 |
0.4 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.1 |
0.5 |
GO:0044330 |
canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 |
0.4 |
GO:0060596 |
mammary placode formation(GO:0060596) |
| 0.1 |
0.8 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.1 |
0.4 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 |
0.1 |
GO:1904058 |
positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 |
0.5 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 |
0.9 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.4 |
GO:0006065 |
UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 |
0.4 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 |
0.1 |
GO:0021852 |
pyramidal neuron migration(GO:0021852) |
| 0.1 |
0.6 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
0.4 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 |
0.5 |
GO:2000313 |
fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 |
0.5 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 |
0.4 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.1 |
0.5 |
GO:0048341 |
paraxial mesoderm formation(GO:0048341) |
| 0.1 |
0.1 |
GO:1901897 |
regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 |
0.7 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.1 |
0.4 |
GO:0001927 |
exocyst assembly(GO:0001927) |
| 0.1 |
0.6 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.2 |
GO:0099601 |
regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.1 |
0.4 |
GO:0030222 |
eosinophil differentiation(GO:0030222) |
| 0.1 |
2.5 |
GO:0048169 |
regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 |
0.7 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 |
0.5 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 |
2.1 |
GO:0010669 |
epithelial structure maintenance(GO:0010669) |
| 0.1 |
0.2 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 |
0.2 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
1.9 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.1 |
0.9 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.1 |
0.3 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) regulation of female receptivity(GO:0045924) |
| 0.1 |
0.5 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 |
0.9 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.8 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
1.6 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.7 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.1 |
0.8 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 |
1.3 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 |
0.9 |
GO:0007213 |
G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 |
0.6 |
GO:0010025 |
wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 |
0.3 |
GO:1903244 |
regulation of cardiac muscle adaptation(GO:0010612) positive regulation of cardiac muscle adaptation(GO:0010615) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 |
0.5 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.1 |
1.7 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 |
0.1 |
GO:0003162 |
atrioventricular node development(GO:0003162) |
| 0.1 |
0.2 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
0.1 |
GO:1900125 |
regulation of hyaluronan biosynthetic process(GO:1900125) |
| 0.1 |
0.2 |
GO:0061418 |
regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 |
0.3 |
GO:0071205 |
protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 |
0.2 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.1 |
0.4 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.1 |
0.9 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.1 |
0.3 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) |
| 0.1 |
0.5 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.1 |
0.9 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 |
0.2 |
GO:0070253 |
somatostatin secretion(GO:0070253) |
| 0.1 |
0.7 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.3 |
GO:0031086 |
nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 |
0.2 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.1 |
0.2 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.1 |
0.8 |
GO:0021756 |
striatum development(GO:0021756) |
| 0.1 |
0.1 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 |
0.5 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 |
1.4 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.1 |
0.3 |
GO:1901421 |
positive regulation of response to alcohol(GO:1901421) |
| 0.1 |
0.6 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 |
0.3 |
GO:1990481 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 |
0.5 |
GO:0044090 |
positive regulation of vacuole organization(GO:0044090) |
| 0.1 |
1.2 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.1 |
0.6 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 |
1.0 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 |
1.5 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.1 |
0.6 |
GO:1902950 |
regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.1 |
0.9 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.1 |
0.8 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 |
0.5 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 |
0.7 |
GO:0000467 |
exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 |
1.0 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.1 |
0.5 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.1 |
0.8 |
GO:0007274 |
neuromuscular synaptic transmission(GO:0007274) |
| 0.1 |
0.3 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 |
0.3 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.1 |
0.3 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 |
0.6 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.1 |
0.6 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 |
0.3 |
GO:0048382 |
neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) mesendoderm development(GO:0048382) |
| 0.1 |
0.7 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 |
2.0 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.1 |
0.2 |
GO:0036302 |
atrioventricular canal development(GO:0036302) |
| 0.1 |
0.4 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 |
0.2 |
GO:1903012 |
positive regulation of bone development(GO:1903012) |
| 0.1 |
0.3 |
GO:0010693 |
negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 |
0.5 |
GO:0034379 |
very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 |
0.4 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 |
0.4 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 |
0.4 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 |
0.1 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.1 |
1.3 |
GO:0080182 |
histone H3-K4 trimethylation(GO:0080182) |
| 0.1 |
0.7 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 |
0.6 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 |
0.6 |
GO:0010839 |
negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 |
0.5 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 |
0.3 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.1 |
5.5 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
0.3 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 |
0.3 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.1 |
0.9 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.1 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.1 |
0.2 |
GO:2001274 |
negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 |
3.4 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.1 |
0.3 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 |
0.2 |
GO:2000832 |
negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 |
0.2 |
GO:0051890 |
regulation of cardioblast differentiation(GO:0051890) |
| 0.1 |
0.2 |
GO:2000819 |
regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 |
0.5 |
GO:0097369 |
sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
2.3 |
GO:0000002 |
mitochondrial genome maintenance(GO:0000002) |
| 0.1 |
0.3 |
GO:0097343 |
ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 |
0.2 |
GO:0014029 |
neural crest formation(GO:0014029) |
| 0.1 |
0.1 |
GO:0097477 |
lateral motor column neuron migration(GO:0097477) |
| 0.1 |
0.4 |
GO:0061622 |
glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 |
0.5 |
GO:0089711 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) L-glutamate transmembrane transport(GO:0089711) |
| 0.1 |
0.7 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.1 |
1.3 |
GO:0051101 |
regulation of DNA binding(GO:0051101) |
| 0.1 |
0.8 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 |
0.6 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 |
0.5 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 |
0.3 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 |
0.2 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.1 |
0.2 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 |
0.5 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.1 |
0.2 |
GO:1901491 |
negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 |
0.3 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 |
0.4 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 |
0.2 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.1 |
0.2 |
GO:0032916 |
transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.1 |
0.4 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.1 |
0.2 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.1 |
0.4 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 |
1.7 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 |
0.3 |
GO:1902224 |
cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.1 |
1.1 |
GO:0009134 |
nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 |
0.3 |
GO:0043987 |
histone H3-S10 phosphorylation(GO:0043987) |
| 0.1 |
0.3 |
GO:2001023 |
regulation of response to drug(GO:2001023) |
| 0.1 |
0.3 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 |
0.8 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 |
0.3 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.1 |
0.9 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 |
0.3 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.1 |
0.3 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.1 |
0.4 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 |
0.3 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
0.2 |
GO:1902774 |
late endosome to lysosome transport(GO:1902774) |
| 0.1 |
0.1 |
GO:0003415 |
chondrocyte hypertrophy(GO:0003415) |
| 0.1 |
0.3 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.1 |
0.2 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 |
0.3 |
GO:0031033 |
myosin filament organization(GO:0031033) |
| 0.1 |
0.3 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 |
1.0 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.1 |
0.1 |
GO:2000015 |
regulation of determination of dorsal identity(GO:2000015) |
| 0.1 |
0.3 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.1 |
0.2 |
GO:0043313 |
regulation of neutrophil degranulation(GO:0043313) |
| 0.1 |
0.4 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 |
4.1 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.1 |
1.4 |
GO:2000272 |
negative regulation of receptor activity(GO:2000272) |
| 0.1 |
0.4 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 |
1.6 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.1 |
1.9 |
GO:0019933 |
cAMP-mediated signaling(GO:0019933) |
| 0.1 |
0.4 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.1 |
0.3 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.1 |
0.4 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 |
0.2 |
GO:2001270 |
regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 |
0.2 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.1 |
0.3 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.6 |
GO:0021952 |
central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 |
0.3 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.1 |
0.7 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 |
1.3 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |
| 0.1 |
0.2 |
GO:0034384 |
high-density lipoprotein particle clearance(GO:0034384) |
| 0.1 |
1.0 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.1 |
0.2 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 |
1.6 |
GO:0007189 |
adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 |
0.2 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.1 |
0.2 |
GO:2000096 |
regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 |
0.2 |
GO:0019405 |
alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 |
0.2 |
GO:0036120 |
response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 |
1.3 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.1 |
0.4 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 |
0.3 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.1 |
GO:0002572 |
pro-T cell differentiation(GO:0002572) |
| 0.1 |
0.3 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.1 |
0.2 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.1 |
0.2 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 |
0.4 |
GO:2000641 |
regulation of early endosome to late endosome transport(GO:2000641) |
| 0.1 |
0.2 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 |
1.0 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 |
0.1 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 |
0.2 |
GO:0060696 |
regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 |
0.2 |
GO:0001778 |
plasma membrane repair(GO:0001778) regulation of glucagon secretion(GO:0070092) |
| 0.0 |
0.2 |
GO:0060690 |
epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.0 |
0.6 |
GO:0060384 |
innervation(GO:0060384) |
| 0.0 |
0.1 |
GO:0003347 |
epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 |
0.1 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.0 |
0.3 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.0 |
0.4 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.2 |
GO:0007603 |
phototransduction, visible light(GO:0007603) |
| 0.0 |
0.2 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 |
0.2 |
GO:1903069 |
regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 |
1.1 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.0 |
GO:0008078 |
mesodermal cell migration(GO:0008078) |
| 0.0 |
0.2 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) |
| 0.0 |
0.2 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.0 |
1.0 |
GO:1902259 |
regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 |
0.2 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.8 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.1 |
GO:0070844 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 |
0.1 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 |
0.2 |
GO:0072365 |
regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 |
0.4 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 |
0.6 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.1 |
GO:2000110 |
negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 |
0.2 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.5 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.8 |
GO:0061001 |
regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 |
0.2 |
GO:0006855 |
drug transmembrane transport(GO:0006855) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 |
0.3 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 |
0.2 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 |
0.3 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.0 |
0.2 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.1 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.0 |
0.6 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
0.4 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 |
1.1 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.2 |
GO:0051238 |
sequestering of calcium ion(GO:0051208) sequestering of metal ion(GO:0051238) regulation of sequestering of calcium ion(GO:0051282) |
| 0.0 |
0.1 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 |
0.2 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.0 |
0.2 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.0 |
0.9 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 |
0.3 |
GO:0030813 |
positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 |
0.3 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.1 |
GO:0046381 |
CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.0 |
0.1 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 |
0.1 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.1 |
GO:0060997 |
dendritic spine morphogenesis(GO:0060997) |
| 0.0 |
0.2 |
GO:0010764 |
negative regulation of fibroblast migration(GO:0010764) |
| 0.0 |
0.1 |
GO:2000273 |
positive regulation of receptor activity(GO:2000273) |
| 0.0 |
0.2 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.0 |
0.1 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.0 |
0.3 |
GO:0030948 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 |
1.7 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.0 |
0.1 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 |
0.2 |
GO:0010747 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 |
0.3 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.0 |
0.9 |
GO:0030819 |
positive regulation of cyclic nucleotide biosynthetic process(GO:0030804) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 |
0.2 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 |
1.2 |
GO:0006220 |
pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.0 |
0.3 |
GO:2000479 |
regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.3 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
0.2 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.0 |
0.1 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.0 |
0.0 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 |
0.2 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.1 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.0 |
0.2 |
GO:1901841 |
regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 |
1.1 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.1 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.0 |
4.6 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.7 |
GO:0034340 |
response to type I interferon(GO:0034340) |
| 0.0 |
0.3 |
GO:0007416 |
synapse assembly(GO:0007416) |
| 0.0 |
0.1 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 |
0.5 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.1 |
GO:0035561 |
regulation of chromatin binding(GO:0035561) |
| 0.0 |
0.4 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.1 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 |
0.4 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
0.2 |
GO:0061000 |
negative regulation of dendritic spine development(GO:0061000) |
| 0.0 |
0.1 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.0 |
0.1 |
GO:0009886 |
post-embryonic morphogenesis(GO:0009886) |
| 0.0 |
0.1 |
GO:0036216 |
response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 |
0.3 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
1.0 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
0.1 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.4 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.5 |
GO:0060292 |
long term synaptic depression(GO:0060292) |
| 0.0 |
0.5 |
GO:0033120 |
positive regulation of RNA splicing(GO:0033120) |
| 0.0 |
0.1 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.0 |
0.1 |
GO:0035898 |
parathyroid hormone secretion(GO:0035898) |
| 0.0 |
0.5 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.2 |
GO:0007183 |
SMAD protein complex assembly(GO:0007183) |
| 0.0 |
0.0 |
GO:0072553 |
terminal button organization(GO:0072553) |
| 0.0 |
0.4 |
GO:0051290 |
protein heterotetramerization(GO:0051290) |
| 0.0 |
0.2 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.2 |
GO:0061298 |
retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 |
1.5 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.5 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.7 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.3 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.9 |
GO:0047496 |
vesicle transport along microtubule(GO:0047496) |
| 0.0 |
0.2 |
GO:0070070 |
proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 |
0.2 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.0 |
0.1 |
GO:0060123 |
regulation of growth hormone secretion(GO:0060123) |
| 0.0 |
0.1 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 |
0.2 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.3 |
GO:0050807 |
regulation of synapse organization(GO:0050807) |
| 0.0 |
0.3 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.0 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.0 |
0.1 |
GO:0090365 |
regulation of mRNA modification(GO:0090365) |
| 0.0 |
0.1 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 |
0.1 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.0 |
0.2 |
GO:0090308 |
regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 |
0.3 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.1 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.4 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.1 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
0.1 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.0 |
0.1 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.0 |
0.7 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.1 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 |
0.1 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.0 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.0 |
0.2 |
GO:0002097 |
tRNA wobble base modification(GO:0002097) |
| 0.0 |
0.3 |
GO:0033572 |
transferrin transport(GO:0033572) |
| 0.0 |
0.1 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.6 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.0 |
0.1 |
GO:0043173 |
purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) nucleotide salvage(GO:0043173) |
| 0.0 |
0.2 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.0 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.0 |
0.1 |
GO:0042059 |
negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.2 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 |
0.2 |
GO:0033275 |
actin-myosin filament sliding(GO:0033275) |
| 0.0 |
0.1 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.0 |
0.2 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.0 |
0.8 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.3 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.1 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 |
0.6 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.5 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
1.4 |
GO:0048813 |
dendrite morphogenesis(GO:0048813) |
| 0.0 |
0.1 |
GO:0000963 |
mitochondrial RNA processing(GO:0000963) |
| 0.0 |
0.2 |
GO:0031103 |
axon regeneration(GO:0031103) |
| 0.0 |
0.1 |
GO:1902837 |
amino acid import into cell(GO:1902837) |
| 0.0 |
0.1 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.0 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.0 |
0.1 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.0 |
0.1 |
GO:0042984 |
amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 |
0.1 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 |
0.1 |
GO:0002666 |
positive regulation of T cell tolerance induction(GO:0002666) immune response-inhibiting signal transduction(GO:0002765) |
| 0.0 |
0.0 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.0 |
0.1 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.1 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.0 |
0.1 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 |
0.1 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.0 |
0.1 |
GO:0009247 |
glycolipid biosynthetic process(GO:0009247) |
| 0.0 |
0.9 |
GO:0043507 |
positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 |
0.1 |
GO:1903624 |
regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.0 |
1.5 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.0 |
0.1 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 |
0.1 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 |
0.3 |
GO:0048599 |
oocyte development(GO:0048599) |
| 0.0 |
0.3 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 |
0.1 |
GO:1900273 |
positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 |
0.1 |
GO:0036265 |
RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 |
0.1 |
GO:0090141 |
positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 |
0.4 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.7 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.0 |
0.2 |
GO:0051930 |
regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 |
0.1 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.2 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.2 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.1 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.0 |
0.0 |
GO:0071636 |
positive regulation of transforming growth factor beta production(GO:0071636) |
| 0.0 |
0.1 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
0.1 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.1 |
GO:0051968 |
positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 |
0.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.1 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.0 |
0.3 |
GO:0016239 |
positive regulation of macroautophagy(GO:0016239) positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.0 |
0.1 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 |
0.1 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 |
0.1 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.0 |
0.1 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.2 |
GO:1902108 |
regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 |
0.2 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.1 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.2 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.1 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.2 |
GO:0034243 |
regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 |
0.0 |
GO:2000612 |
positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid hormone generation(GO:2000609) positive regulation of thyroid hormone generation(GO:2000611) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 |
0.2 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 |
0.1 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.3 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.1 |
GO:0071493 |
cellular response to UV-B(GO:0071493) |
| 0.0 |
0.0 |
GO:1900533 |
medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 |
0.6 |
GO:0050709 |
negative regulation of protein secretion(GO:0050709) |
| 0.0 |
0.2 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 |
0.0 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 |
0.1 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 |
0.2 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
0.0 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.0 |
GO:0045346 |
regulation of MHC class II biosynthetic process(GO:0045346) negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 |
0.1 |
GO:0014046 |
dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 |
0.0 |
GO:0071220 |
response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.0 |
0.1 |
GO:0072384 |
organelle transport along microtubule(GO:0072384) |