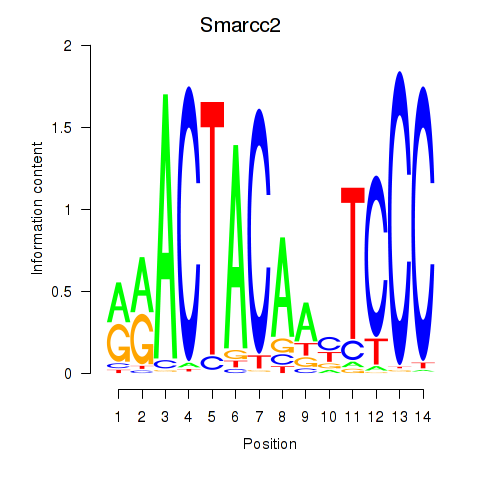
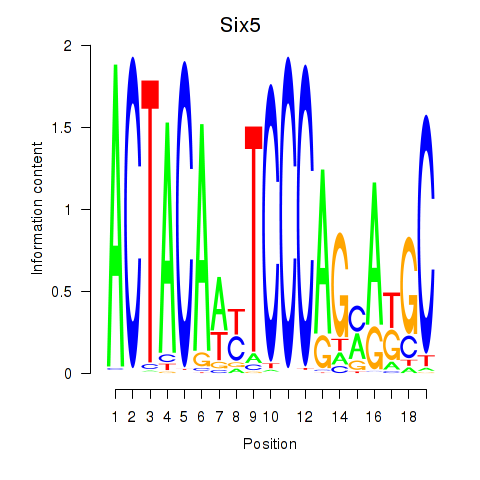
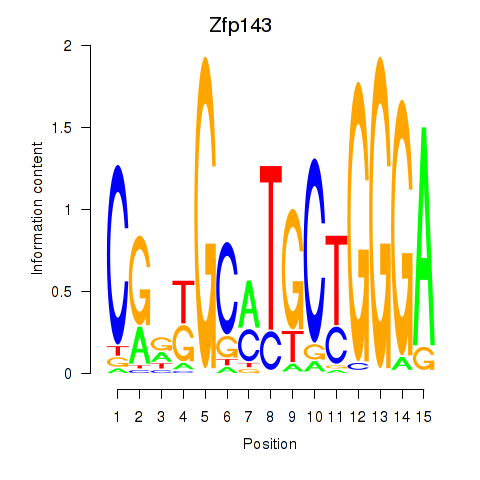
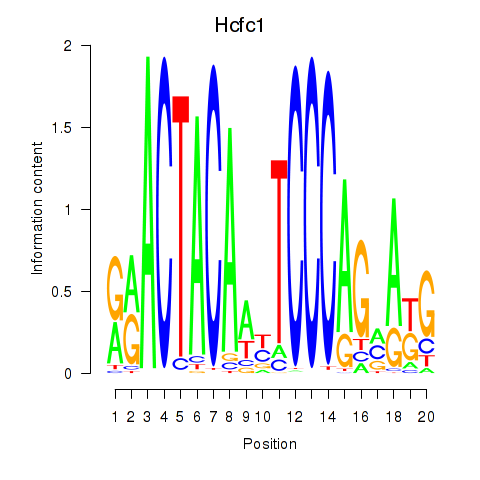
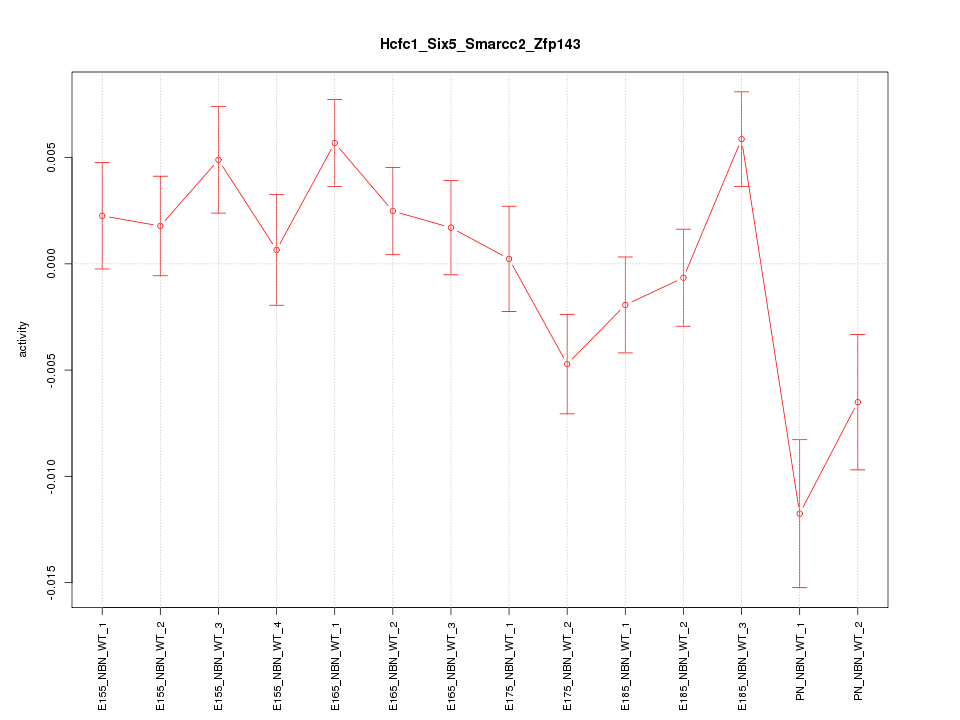
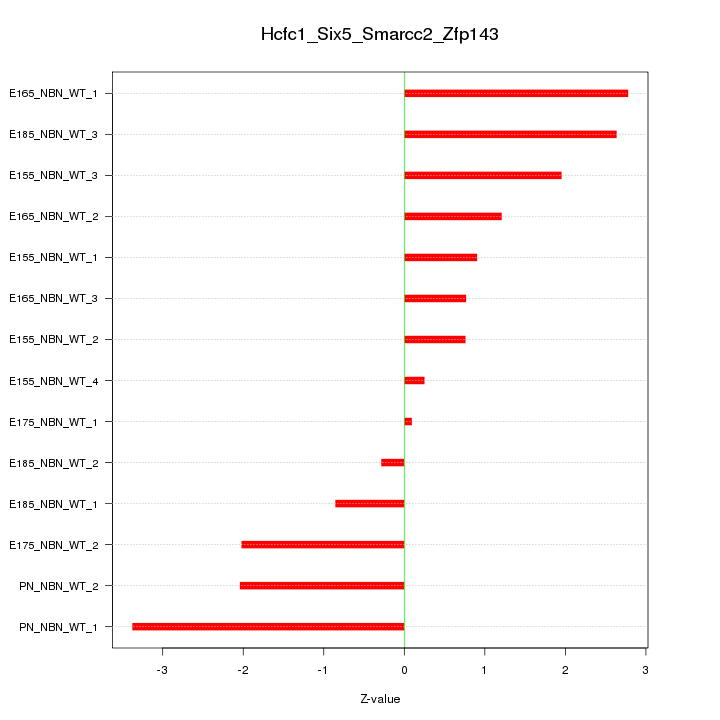
| 0.7 |
2.0 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.5 |
0.5 |
GO:0032240 |
negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.5 |
2.5 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.3 |
0.3 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.3 |
1.9 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.3 |
0.9 |
GO:1903896 |
positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.3 |
0.9 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.3 |
1.1 |
GO:0045343 |
MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.3 |
0.8 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 |
0.6 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.2 |
0.6 |
GO:0000454 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 |
0.6 |
GO:0034968 |
histone lysine methylation(GO:0034968) |
| 0.2 |
1.3 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.2 |
0.5 |
GO:0046381 |
CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.2 |
2.2 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.2 |
0.5 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.2 |
0.6 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.2 |
0.5 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.2 |
0.5 |
GO:0035552 |
oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.2 |
0.8 |
GO:0060334 |
regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 |
0.4 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 |
1.6 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 |
1.5 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.1 |
0.4 |
GO:0046168 |
glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 |
0.4 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.1 |
0.7 |
GO:0009642 |
response to light intensity(GO:0009642) |
| 0.1 |
0.7 |
GO:0035247 |
peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.1 |
1.0 |
GO:0032308 |
positive regulation of prostaglandin secretion(GO:0032308) |
| 0.1 |
0.3 |
GO:0000350 |
generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 |
0.8 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.1 |
1.1 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 |
0.6 |
GO:0030423 |
targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 |
0.7 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.1 |
0.4 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.1 |
1.1 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 |
0.7 |
GO:0061052 |
negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 |
0.6 |
GO:0036508 |
protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.1 |
0.6 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.1 |
0.6 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 |
0.6 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) regulation of stem cell division(GO:2000035) |
| 0.1 |
0.7 |
GO:0016078 |
tRNA catabolic process(GO:0016078) |
| 0.1 |
0.4 |
GO:0061511 |
centriole elongation(GO:0061511) |
| 0.1 |
0.5 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 |
2.1 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 |
0.6 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.1 |
0.7 |
GO:0048341 |
paraxial mesoderm formation(GO:0048341) |
| 0.1 |
0.3 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 |
1.3 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 |
0.3 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 |
0.5 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 |
1.6 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.1 |
0.3 |
GO:1901096 |
regulation of autophagosome maturation(GO:1901096) |
| 0.1 |
0.4 |
GO:0006659 |
phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 |
0.6 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 |
0.4 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 |
0.5 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.1 |
0.3 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.1 |
0.2 |
GO:0009838 |
abscission(GO:0009838) |
| 0.1 |
0.2 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.1 |
0.3 |
GO:1903265 |
keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 |
0.2 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.1 |
0.3 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 |
0.6 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.1 |
0.3 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 |
0.5 |
GO:0070417 |
cellular response to cold(GO:0070417) |
| 0.1 |
1.9 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 |
0.6 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.1 |
0.5 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.1 |
1.0 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 |
0.3 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 |
0.5 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 |
0.6 |
GO:1900118 |
negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.1 |
0.4 |
GO:0036112 |
medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 |
0.2 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 |
0.1 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 |
0.9 |
GO:0070131 |
positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 |
0.7 |
GO:0035090 |
maintenance of apical/basal cell polarity(GO:0035090) |
| 0.1 |
0.6 |
GO:0034351 |
negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 |
0.1 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.1 |
1.2 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.1 |
0.6 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.1 |
0.4 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 |
0.3 |
GO:2001270 |
regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.1 |
1.0 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.1 |
0.3 |
GO:0006481 |
C-terminal protein methylation(GO:0006481) |
| 0.1 |
1.1 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.1 |
0.4 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 |
0.1 |
GO:0050689 |
negative regulation of defense response to virus by host(GO:0050689) |
| 0.1 |
0.2 |
GO:0035928 |
RNA import into mitochondrion(GO:0035927) rRNA import into mitochondrion(GO:0035928) |
| 0.1 |
0.4 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.1 |
0.5 |
GO:0044539 |
long-chain fatty acid import(GO:0044539) |
| 0.1 |
0.2 |
GO:0071609 |
neutrophil mediated killing of bacterium(GO:0070944) chemokine (C-C motif) ligand 5 production(GO:0071609) |
| 0.1 |
0.2 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.1 |
0.8 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 |
0.3 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 |
0.1 |
GO:0009193 |
pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 |
0.2 |
GO:0030647 |
polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.1 |
0.4 |
GO:0090503 |
RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 |
0.2 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 |
0.6 |
GO:0030049 |
muscle filament sliding(GO:0030049) |
| 0.1 |
0.2 |
GO:0010626 |
negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 |
0.9 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 |
0.2 |
GO:0015959 |
diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.1 |
0.7 |
GO:0010807 |
regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 |
0.6 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 |
0.7 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.1 |
0.2 |
GO:0010793 |
regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.1 |
0.9 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 |
0.2 |
GO:0042776 |
mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 |
0.6 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 |
0.3 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 |
0.6 |
GO:0052646 |
alditol phosphate metabolic process(GO:0052646) |
| 0.1 |
0.2 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 |
2.1 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.1 |
0.2 |
GO:1901301 |
regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 |
0.2 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 |
0.2 |
GO:0060912 |
cardiac cell fate specification(GO:0060912) |
| 0.1 |
0.4 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.0 |
0.4 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.1 |
GO:1901421 |
positive regulation of response to alcohol(GO:1901421) |
| 0.0 |
0.3 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 |
0.1 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.0 |
0.7 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.2 |
GO:2000676 |
positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 |
0.1 |
GO:0048611 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 |
1.1 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.0 |
0.3 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.6 |
GO:0032506 |
cytokinetic process(GO:0032506) |
| 0.0 |
0.2 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.0 |
0.1 |
GO:0034140 |
negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) aggrephagy(GO:0035973) |
| 0.0 |
0.1 |
GO:0048239 |
negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 |
0.1 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.0 |
0.4 |
GO:0002544 |
chronic inflammatory response(GO:0002544) |
| 0.0 |
0.0 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 |
0.3 |
GO:0019370 |
leukotriene biosynthetic process(GO:0019370) |
| 0.0 |
0.2 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.6 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.2 |
GO:0008298 |
intracellular mRNA localization(GO:0008298) |
| 0.0 |
0.2 |
GO:1901069 |
guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 |
0.4 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 |
1.2 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.4 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 |
0.3 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.1 |
GO:0046959 |
habituation(GO:0046959) |
| 0.0 |
0.2 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 |
0.1 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) |
| 0.0 |
0.1 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 |
0.2 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.7 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.1 |
GO:2000304 |
positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 |
0.1 |
GO:0045911 |
positive regulation of DNA recombination(GO:0045911) |
| 0.0 |
0.1 |
GO:0006549 |
isoleucine metabolic process(GO:0006549) |
| 0.0 |
2.8 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.0 |
0.1 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.0 |
0.6 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
0.2 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.0 |
0.1 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 |
1.2 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
0.1 |
GO:0002922 |
positive regulation of humoral immune response(GO:0002922) |
| 0.0 |
0.1 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.0 |
0.2 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.4 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.0 |
0.2 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.0 |
0.2 |
GO:0016114 |
terpenoid biosynthetic process(GO:0016114) |
| 0.0 |
0.1 |
GO:0003245 |
growth involved in heart morphogenesis(GO:0003241) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 |
0.1 |
GO:0030300 |
regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.0 |
0.1 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.0 |
0.2 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.1 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.0 |
0.6 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
0.2 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.0 |
1.0 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.4 |
GO:0046929 |
negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.0 |
0.6 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.1 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 |
0.3 |
GO:0060575 |
intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 |
0.2 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 |
0.1 |
GO:0018008 |
N-terminal peptidyl-glycine N-myristoylation(GO:0018008) peptidyl-glycine modification(GO:0018201) |
| 0.0 |
0.2 |
GO:0015919 |
peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 |
0.2 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.0 |
0.2 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.3 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.2 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.1 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 |
0.6 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 |
0.2 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.3 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.1 |
GO:0008655 |
pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 |
0.3 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
0.0 |
GO:1904468 |
negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.0 |
0.4 |
GO:0051383 |
kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.0 |
0.1 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.0 |
0.1 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.0 |
0.2 |
GO:0030174 |
regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 |
0.3 |
GO:0051966 |
regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 |
0.4 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.1 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.0 |
0.1 |
GO:1903204 |
negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 |
2.0 |
GO:0016579 |
protein deubiquitination(GO:0016579) |
| 0.0 |
0.0 |
GO:0032070 |
regulation of deoxyribonuclease activity(GO:0032070) positive regulation of deoxyribonuclease activity(GO:0032077) |
| 0.0 |
0.8 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.0 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 |
0.5 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.2 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 |
0.1 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
1.4 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.3 |
GO:0009191 |
ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 |
0.2 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.1 |
GO:0060065 |
uterus development(GO:0060065) |
| 0.0 |
0.3 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
0.5 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.1 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 |
0.1 |
GO:0061718 |
NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glycolytic process through glucose-1-phosphate(GO:0061622) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 |
0.0 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.0 |
0.1 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 |
0.1 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 |
0.6 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 |
0.9 |
GO:0019933 |
cAMP-mediated signaling(GO:0019933) |
| 0.0 |
0.3 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.1 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 |
1.3 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.5 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.3 |
GO:0043584 |
nose development(GO:0043584) |
| 0.0 |
0.3 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.0 |
0.1 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.0 |
0.1 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.2 |
GO:0045047 |
protein targeting to ER(GO:0045047) |
| 0.0 |
0.2 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.1 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 |
0.0 |
GO:0018197 |
peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 |
0.0 |
GO:0048703 |
embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 |
0.1 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 |
0.1 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 |
0.4 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
0.1 |
GO:0009303 |
rRNA transcription(GO:0009303) |
| 0.0 |
0.2 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.2 |
GO:0007602 |
phototransduction(GO:0007602) |
| 0.0 |
0.2 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 |
0.1 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
0.2 |
GO:0045191 |
regulation of isotype switching(GO:0045191) |
| 0.0 |
0.1 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 |
0.9 |
GO:0090305 |
nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 |
0.0 |
GO:0071560 |
cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 |
0.1 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.0 |
0.1 |
GO:0031145 |
anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 |
0.2 |
GO:0080182 |
histone H3-K4 trimethylation(GO:0080182) |
| 0.0 |
0.1 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.0 |
0.3 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.1 |
GO:0071169 |
establishment of protein localization to chromatin(GO:0071169) |
| 0.0 |
0.2 |
GO:0006103 |
2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 |
0.0 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 |
0.1 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 |
0.2 |
GO:2000009 |
negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 |
0.3 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
1.4 |
GO:0098792 |
xenophagy(GO:0098792) |
| 0.0 |
1.5 |
GO:0008344 |
adult locomotory behavior(GO:0008344) |
| 0.0 |
0.9 |
GO:0016575 |
histone deacetylation(GO:0016575) |
| 0.0 |
0.1 |
GO:0071479 |
cellular response to ionizing radiation(GO:0071479) |
| 0.0 |
0.3 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.1 |
GO:0034312 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.0 |
0.4 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |
| 0.0 |
0.3 |
GO:0048662 |
negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 |
0.0 |
GO:0070257 |
positive regulation of mucus secretion(GO:0070257) |
| 0.0 |
0.0 |
GO:1901187 |
regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 |
0.2 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.3 |
GO:0008214 |
protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 |
0.1 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.8 |
GO:0043407 |
negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 |
0.3 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.0 |
0.2 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.0 |
0.3 |
GO:2001240 |
negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 |
0.1 |
GO:0070126 |
mitochondrial translational termination(GO:0070126) |
| 0.0 |
0.1 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 |
0.0 |
GO:0000720 |
pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 |
0.1 |
GO:0032471 |
negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.0 |
0.4 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.4 |
GO:0045668 |
negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 |
0.0 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 |
0.0 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.0 |
0.1 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.1 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 |
0.1 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 |
0.1 |
GO:0009186 |
deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 |
0.1 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.6 |
GO:0000423 |
macromitophagy(GO:0000423) |
| 0.0 |
1.6 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |