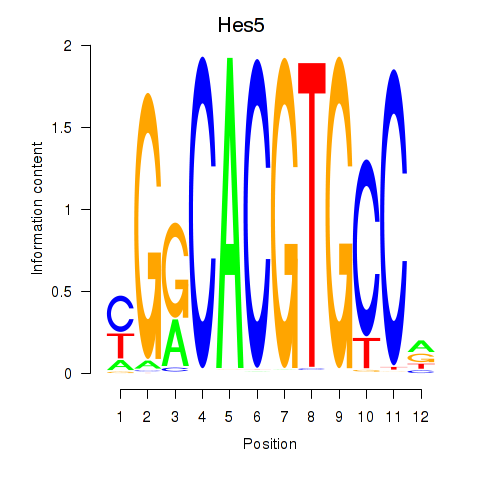
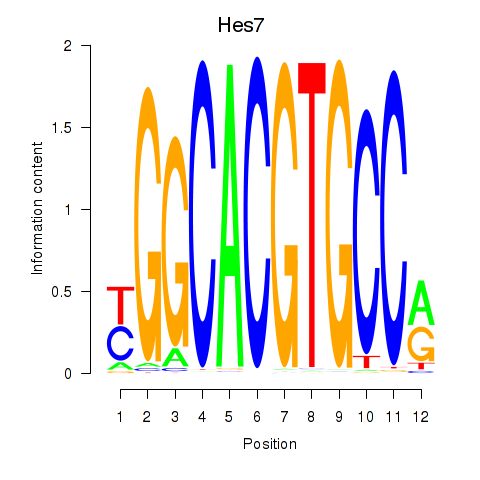
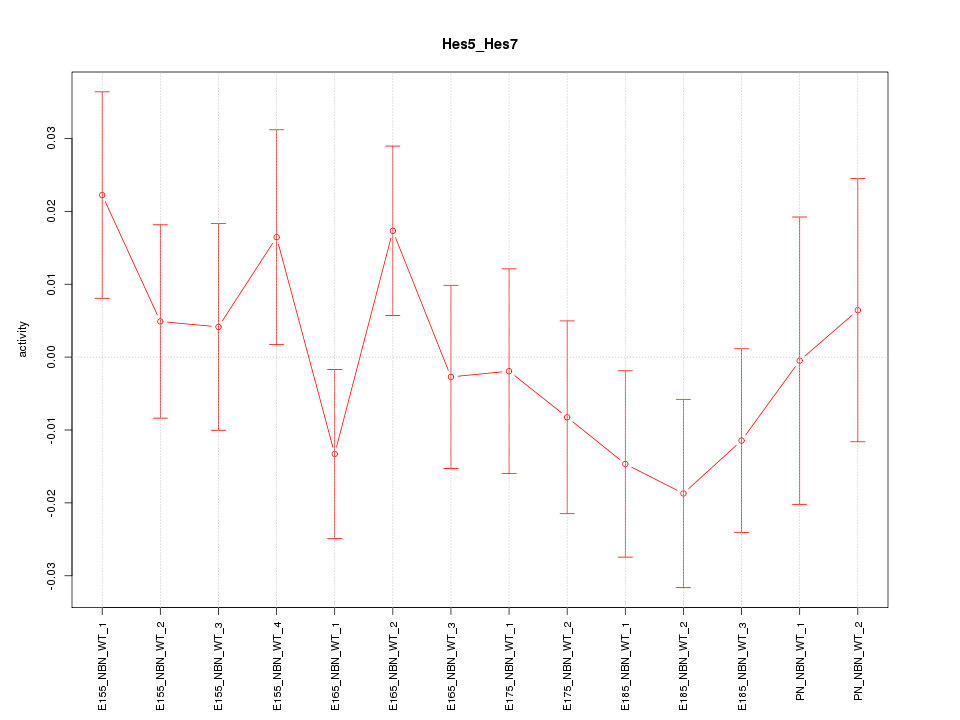
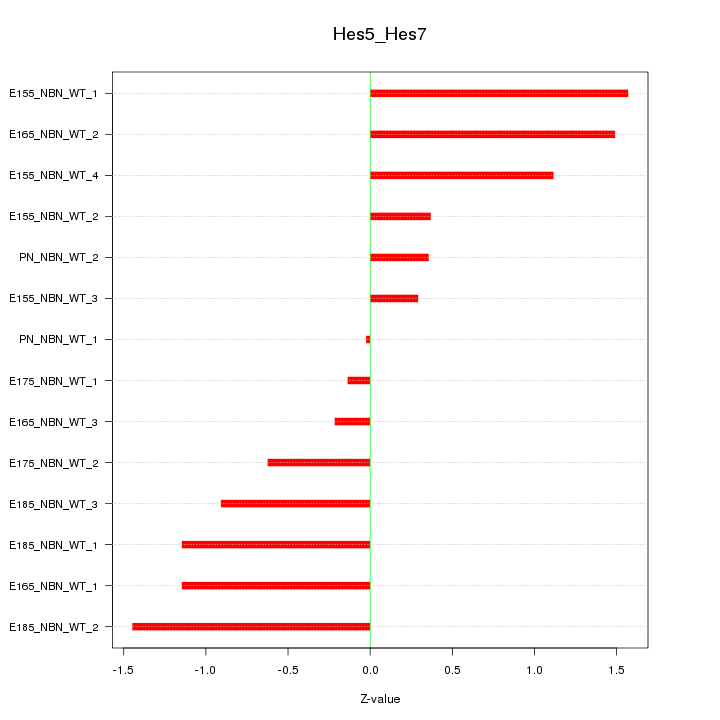
| 1.3 |
12.0 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.4 |
1.1 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.2 |
2.5 |
GO:0042095 |
interferon-gamma biosynthetic process(GO:0042095) |
| 0.2 |
0.7 |
GO:0051771 |
negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 |
0.4 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 |
0.7 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.1 |
0.7 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) |
| 0.1 |
1.1 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.1 |
0.4 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.1 |
0.6 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 |
0.3 |
GO:2000017 |
positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 |
0.3 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 |
1.4 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.2 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.0 |
0.7 |
GO:0045187 |
regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.8 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.2 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 |
0.4 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
1.1 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 |
0.1 |
GO:0010814 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 |
0.3 |
GO:0033690 |
positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 |
0.3 |
GO:0032366 |
intracellular sterol transport(GO:0032366) |
| 0.0 |
0.1 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
0.3 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 |
0.1 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 |
0.3 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.1 |
GO:0019433 |
triglyceride catabolic process(GO:0019433) |
| 0.0 |
0.1 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 |
0.1 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.0 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.1 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.2 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 |
0.1 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 |
0.2 |
GO:0032402 |
melanosome transport(GO:0032402) |
| 0.0 |
0.4 |
GO:0060351 |
cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 |
0.1 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.2 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |