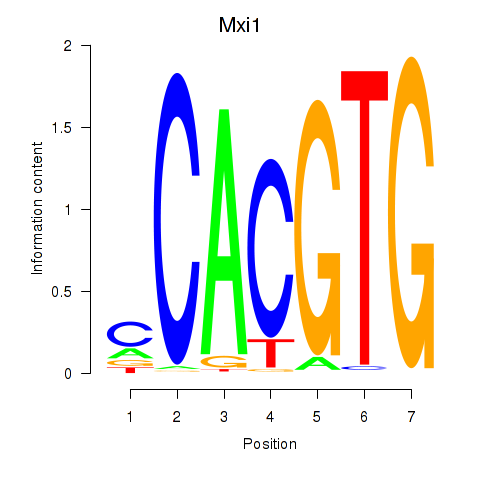
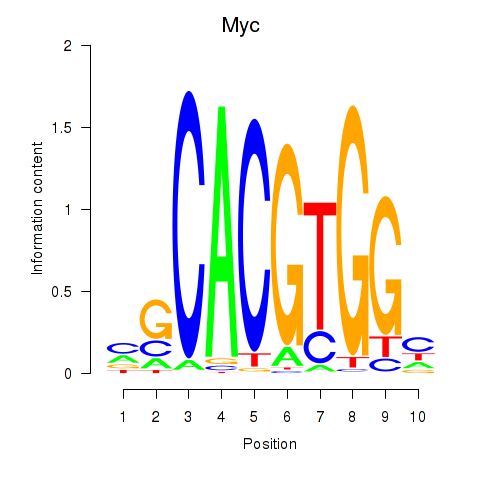
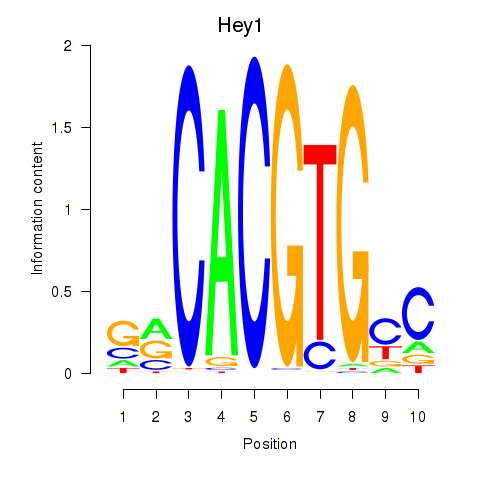
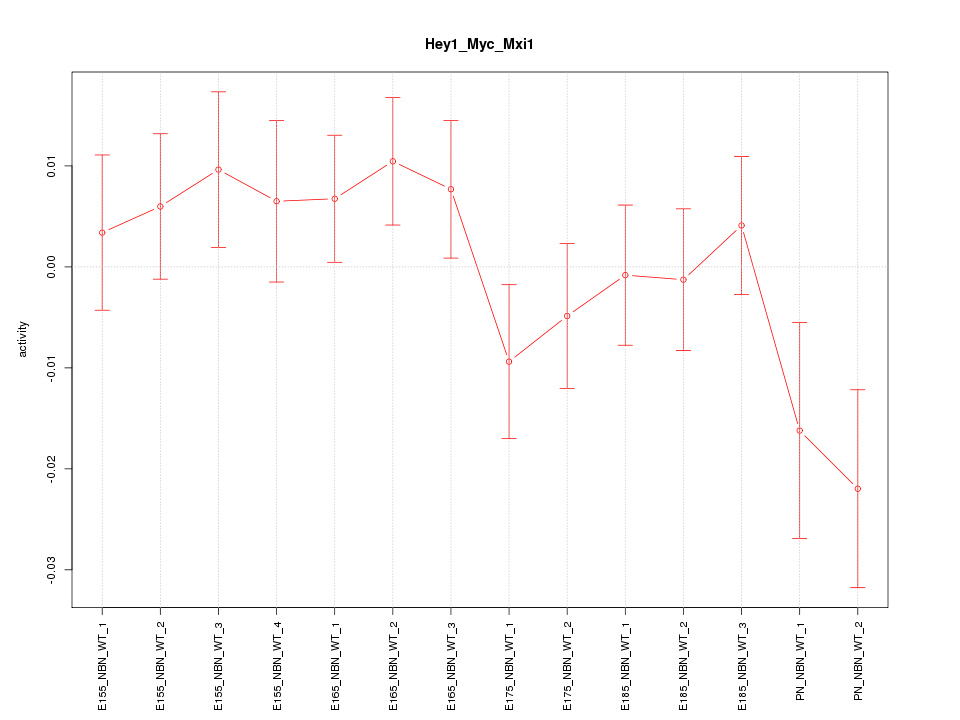
| 2.2 |
15.6 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.4 |
4.2 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 1.2 |
3.7 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.8 |
2.4 |
GO:0036353 |
histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.8 |
3.1 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.8 |
3.9 |
GO:0002121 |
inter-male aggressive behavior(GO:0002121) |
| 0.7 |
2.2 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.7 |
2.0 |
GO:0009644 |
response to high light intensity(GO:0009644) |
| 0.6 |
3.2 |
GO:0006189 |
'de novo' IMP biosynthetic process(GO:0006189) |
| 0.6 |
2.5 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.6 |
1.7 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.5 |
2.2 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.5 |
1.6 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.5 |
2.0 |
GO:0097296 |
activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.5 |
1.5 |
GO:0070428 |
granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.5 |
1.4 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.4 |
2.2 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.4 |
2.0 |
GO:0048631 |
regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.4 |
1.2 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.4 |
2.7 |
GO:0035902 |
response to immobilization stress(GO:0035902) |
| 0.4 |
1.9 |
GO:0035984 |
response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.4 |
2.6 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.4 |
1.1 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.3 |
1.4 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.3 |
3.0 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 |
1.0 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 |
2.6 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 |
0.9 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.3 |
0.9 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) |
| 0.3 |
0.9 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.3 |
1.2 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.3 |
0.6 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.3 |
1.7 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 |
1.4 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.3 |
0.8 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.3 |
1.6 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.3 |
1.1 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.3 |
2.9 |
GO:0006228 |
UTP biosynthetic process(GO:0006228) |
| 0.3 |
0.8 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.3 |
0.5 |
GO:0035973 |
aggrephagy(GO:0035973) |
| 0.3 |
2.3 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 |
0.5 |
GO:2001012 |
mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.2 |
1.2 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 |
3.0 |
GO:0070131 |
positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 |
1.0 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.2 |
0.7 |
GO:0046370 |
fructose biosynthetic process(GO:0046370) |
| 0.2 |
0.5 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 |
0.5 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.2 |
3.3 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.2 |
0.7 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.2 |
2.3 |
GO:0002072 |
optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.2 |
0.7 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.2 |
0.7 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 |
0.4 |
GO:2000468 |
regulation of peroxidase activity(GO:2000468) |
| 0.2 |
0.8 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 |
1.3 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.2 |
0.6 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.2 |
0.6 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 |
1.4 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 |
2.4 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.2 |
1.0 |
GO:1902570 |
protein localization to nucleolus(GO:1902570) |
| 0.2 |
2.2 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 |
1.1 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 |
0.9 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 |
0.7 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.2 |
0.9 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 |
0.5 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.2 |
0.5 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 |
0.9 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 |
0.7 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 |
0.5 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.2 |
1.3 |
GO:0009125 |
nucleoside monophosphate catabolic process(GO:0009125) |
| 0.2 |
1.5 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 |
0.7 |
GO:0030214 |
hyaluronan catabolic process(GO:0030214) |
| 0.2 |
0.5 |
GO:0090298 |
negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.2 |
0.6 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.2 |
0.3 |
GO:0033159 |
negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.2 |
0.2 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.1 |
1.6 |
GO:1901250 |
regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.1 |
1.0 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.1 |
0.6 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 |
0.6 |
GO:0042364 |
water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 |
0.9 |
GO:1902074 |
response to salt(GO:1902074) |
| 0.1 |
0.3 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 |
2.8 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
0.7 |
GO:0070827 |
chromatin maintenance(GO:0070827) |
| 0.1 |
0.4 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.1 |
0.7 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 |
0.6 |
GO:0045343 |
MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 |
1.8 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 |
0.4 |
GO:1904049 |
regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 |
0.5 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.1 |
0.4 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 |
0.4 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 |
1.0 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 |
1.2 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
0.4 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 |
0.5 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 |
2.5 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 |
0.4 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.1 |
0.7 |
GO:0072553 |
terminal button organization(GO:0072553) |
| 0.1 |
0.4 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 |
0.5 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.1 |
1.0 |
GO:0019348 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) dolichol metabolic process(GO:0019348) |
| 0.1 |
0.9 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.1 |
0.6 |
GO:0046684 |
response to insecticide(GO:0017085) response to pyrethroid(GO:0046684) |
| 0.1 |
0.6 |
GO:0006982 |
response to lipid hydroperoxide(GO:0006982) |
| 0.1 |
0.3 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 0.1 |
1.6 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) |
| 0.1 |
0.1 |
GO:1900368 |
regulation of RNA interference(GO:1900368) |
| 0.1 |
0.5 |
GO:0008355 |
olfactory learning(GO:0008355) |
| 0.1 |
1.4 |
GO:0090502 |
RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 |
0.3 |
GO:0038095 |
Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.1 |
1.4 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
0.2 |
GO:1902414 |
protein localization to cell junction(GO:1902414) |
| 0.1 |
0.5 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.1 |
0.4 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 |
0.7 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 |
0.1 |
GO:0019896 |
axonal transport of mitochondrion(GO:0019896) |
| 0.1 |
0.4 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 0.1 |
0.3 |
GO:0060830 |
ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 |
0.3 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 |
0.3 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 |
0.6 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 |
0.3 |
GO:0046881 |
positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 |
0.9 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.1 |
0.8 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.5 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 |
0.5 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.1 |
0.7 |
GO:0006000 |
fructose metabolic process(GO:0006000) |
| 0.1 |
0.2 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 |
0.1 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.4 |
GO:0021730 |
trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.1 |
1.4 |
GO:0097284 |
hepatocyte apoptotic process(GO:0097284) |
| 0.1 |
2.8 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.1 |
0.4 |
GO:0060266 |
positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.1 |
0.7 |
GO:0044351 |
macropinocytosis(GO:0044351) |
| 0.1 |
0.5 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 |
4.7 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.1 |
0.4 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.1 |
2.8 |
GO:0051443 |
positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 |
0.4 |
GO:0071205 |
clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 |
0.6 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
2.3 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 |
0.2 |
GO:0051572 |
negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.1 |
3.0 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.1 |
0.3 |
GO:0040038 |
polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 |
0.9 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 |
0.3 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 |
0.4 |
GO:0071638 |
negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 |
0.5 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.1 |
0.4 |
GO:1902861 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) copper ion import into cell(GO:1902861) |
| 0.1 |
0.3 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.1 |
0.2 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.1 |
0.6 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.1 |
0.4 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 |
0.2 |
GO:0046654 |
tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 |
0.7 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 |
0.2 |
GO:0045869 |
negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.1 |
0.3 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.1 |
1.2 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 |
0.2 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.1 |
0.1 |
GO:1901857 |
positive regulation of cellular respiration(GO:1901857) |
| 0.1 |
0.3 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 |
0.2 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.1 |
0.3 |
GO:0097411 |
hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 |
0.4 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.4 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
1.4 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 |
0.3 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 |
0.2 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.1 |
0.1 |
GO:0006537 |
glutamate biosynthetic process(GO:0006537) |
| 0.1 |
0.9 |
GO:0097688 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 |
0.2 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.1 |
0.6 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 |
0.4 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.1 |
2.0 |
GO:0060351 |
cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.1 |
0.3 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.1 |
1.1 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 |
0.7 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 |
0.1 |
GO:0071880 |
adrenergic receptor signaling pathway(GO:0071875) adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 |
0.8 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.1 |
0.7 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.1 |
0.3 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.1 |
0.4 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 |
0.3 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 |
0.9 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.1 |
0.5 |
GO:0046015 |
regulation of transcription by glucose(GO:0046015) |
| 0.1 |
0.3 |
GO:0032792 |
negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 |
0.2 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.1 |
0.6 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 |
0.3 |
GO:0036265 |
RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 |
0.2 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.1 |
0.3 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 |
0.6 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.1 |
0.1 |
GO:2000323 |
negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 |
0.2 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 |
1.1 |
GO:0097576 |
autophagosome maturation(GO:0097352) vacuole fusion(GO:0097576) |
| 0.1 |
0.1 |
GO:0060481 |
lobar bronchus epithelium development(GO:0060481) |
| 0.1 |
0.8 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.1 |
0.5 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.1 |
0.3 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 |
0.6 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 |
0.3 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 |
0.3 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 |
0.2 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |
| 0.1 |
0.2 |
GO:0031120 |
snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 |
0.3 |
GO:0010944 |
negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 |
0.2 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.1 |
0.2 |
GO:0006659 |
phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 |
0.4 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.1 |
0.1 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.1 |
0.4 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.1 |
0.2 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.1 |
0.2 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.1 |
0.1 |
GO:0070257 |
positive regulation of mucus secretion(GO:0070257) |
| 0.1 |
0.9 |
GO:0006308 |
DNA catabolic process(GO:0006308) |
| 0.1 |
0.2 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 |
0.6 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 |
1.0 |
GO:0018279 |
protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 |
0.3 |
GO:0019249 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.1 |
0.7 |
GO:0090261 |
positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 |
0.3 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 |
0.8 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 |
0.2 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.0 |
0.1 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 |
0.4 |
GO:2000074 |
regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 |
0.7 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.0 |
0.1 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 |
1.0 |
GO:0045672 |
positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 |
0.4 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 |
0.2 |
GO:0018364 |
peptidyl-glutamine methylation(GO:0018364) |
| 0.0 |
0.2 |
GO:0006046 |
N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 |
0.6 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.2 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.0 |
0.1 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.0 |
0.4 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
1.0 |
GO:0010738 |
regulation of protein kinase A signaling(GO:0010738) |
| 0.0 |
0.2 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 |
0.2 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.3 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
1.1 |
GO:0030488 |
tRNA methylation(GO:0030488) |
| 0.0 |
0.7 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.4 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.2 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.0 |
0.6 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.0 |
0.3 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 |
0.2 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 |
0.2 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.0 |
0.1 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.0 |
0.2 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.0 |
0.2 |
GO:0042699 |
follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 |
0.4 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.2 |
GO:0045072 |
regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 |
0.2 |
GO:0009098 |
branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 |
0.5 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 |
1.3 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.8 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.2 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 |
0.7 |
GO:0051895 |
negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 |
0.2 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.2 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.3 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.0 |
0.2 |
GO:0060510 |
Type II pneumocyte differentiation(GO:0060510) |
| 0.0 |
0.3 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 |
0.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.2 |
GO:0008655 |
pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 |
0.1 |
GO:0032439 |
regulation of cholesterol esterification(GO:0010872) endosome localization(GO:0032439) |
| 0.0 |
1.1 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:0090526 |
glucosamine metabolic process(GO:0006041) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 |
0.5 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.0 |
0.3 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
1.6 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.1 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 |
0.5 |
GO:0072600 |
establishment of protein localization to Golgi(GO:0072600) |
| 0.0 |
2.2 |
GO:0006405 |
RNA export from nucleus(GO:0006405) |
| 0.0 |
0.3 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.0 |
0.6 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.4 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
1.8 |
GO:0051453 |
regulation of intracellular pH(GO:0051453) |
| 0.0 |
1.0 |
GO:0051291 |
protein heterooligomerization(GO:0051291) |
| 0.0 |
0.2 |
GO:0008089 |
anterograde axonal transport(GO:0008089) |
| 0.0 |
0.1 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.0 |
0.8 |
GO:0009268 |
response to pH(GO:0009268) |
| 0.0 |
0.2 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 |
0.5 |
GO:0090140 |
regulation of mitochondrial fission(GO:0090140) |
| 0.0 |
0.3 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.0 |
0.9 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
0.7 |
GO:0040015 |
negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 |
0.2 |
GO:0018195 |
peptidyl-arginine modification(GO:0018195) |
| 0.0 |
0.2 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.1 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.0 |
0.6 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
0.2 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 |
0.1 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 |
0.2 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 |
0.4 |
GO:0006930 |
substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 |
0.4 |
GO:0043457 |
regulation of cellular respiration(GO:0043457) |
| 0.0 |
0.6 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.1 |
GO:0000957 |
mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 |
0.3 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.0 |
0.1 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.0 |
0.1 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 |
0.0 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) |
| 0.0 |
0.1 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 |
0.4 |
GO:0031987 |
locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 |
0.1 |
GO:0014028 |
notochord formation(GO:0014028) |
| 0.0 |
0.1 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 |
0.2 |
GO:0003056 |
regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 |
0.3 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.0 |
0.1 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.0 |
0.1 |
GO:0032053 |
ciliary basal body organization(GO:0032053) |
| 0.0 |
0.3 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.1 |
GO:0019478 |
D-amino acid catabolic process(GO:0019478) |
| 0.0 |
0.5 |
GO:0071391 |
cellular response to estrogen stimulus(GO:0071391) |
| 0.0 |
0.2 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 |
0.2 |
GO:0048009 |
insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 |
0.3 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 |
0.4 |
GO:0051654 |
establishment of mitochondrion localization(GO:0051654) |
| 0.0 |
0.4 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
0.2 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 |
0.1 |
GO:0045351 |
type I interferon biosynthetic process(GO:0045351) |
| 0.0 |
0.5 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.0 |
0.1 |
GO:0031133 |
regulation of axon diameter(GO:0031133) |
| 0.0 |
0.2 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.3 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.1 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.2 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 |
0.8 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.1 |
GO:0060287 |
epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 |
1.7 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.5 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.2 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.0 |
0.2 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
0.0 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.0 |
0.5 |
GO:0035774 |
positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 |
0.4 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.0 |
GO:1902224 |
cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.0 |
0.8 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.2 |
GO:0043206 |
extracellular fibril organization(GO:0043206) |
| 0.0 |
0.2 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 |
0.1 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 |
0.1 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 |
0.1 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 |
0.1 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 |
0.1 |
GO:0060346 |
bone trabecula formation(GO:0060346) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.0 |
0.1 |
GO:0046502 |
uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 |
1.9 |
GO:2001237 |
negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 |
0.0 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 |
0.6 |
GO:0086010 |
membrane depolarization during action potential(GO:0086010) |
| 0.0 |
0.4 |
GO:0003009 |
skeletal muscle contraction(GO:0003009) |
| 0.0 |
0.3 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.2 |
GO:0002052 |
positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 |
0.1 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.0 |
1.2 |
GO:0006892 |
post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 |
0.1 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 |
0.2 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.0 |
0.1 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 |
0.4 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
2.2 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 |
0.0 |
GO:0000720 |
pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 |
0.9 |
GO:1902850 |
mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 |
0.2 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.3 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.0 |
0.1 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.2 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.2 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.2 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
0.1 |
GO:0032402 |
establishment of melanosome localization(GO:0032401) melanosome transport(GO:0032402) |
| 0.0 |
0.5 |
GO:0006024 |
glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 |
0.3 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.3 |
GO:0006101 |
citrate metabolic process(GO:0006101) |
| 0.0 |
0.3 |
GO:0048148 |
behavioral response to cocaine(GO:0048148) |
| 0.0 |
0.1 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 |
0.3 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.0 |
0.3 |
GO:0040019 |
positive regulation of embryonic development(GO:0040019) |
| 0.0 |
0.5 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.2 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 |
0.3 |
GO:0006582 |
melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 |
0.3 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.0 |
0.1 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.0 |
0.2 |
GO:0022401 |
desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.0 |
0.2 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 |
0.1 |
GO:0046166 |
glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 |
0.5 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.8 |
GO:0090305 |
nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 |
0.2 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.0 |
0.3 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 |
0.3 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.0 |
0.7 |
GO:0006487 |
protein N-linked glycosylation(GO:0006487) |
| 0.0 |
0.1 |
GO:0071426 |
ribonucleoprotein complex export from nucleus(GO:0071426) |
| 0.0 |
0.4 |
GO:0006220 |
pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.0 |
0.1 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.1 |
GO:0060122 |
inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 |
0.0 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.0 |
0.4 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.5 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.1 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.1 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.0 |
1.4 |
GO:0030705 |
cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 |
0.1 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 |
0.6 |
GO:0007569 |
cell aging(GO:0007569) |
| 0.0 |
0.0 |
GO:0043046 |
DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 |
0.2 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 |
0.1 |
GO:0051341 |
regulation of oxidoreductase activity(GO:0051341) |
| 0.0 |
0.1 |
GO:0021554 |
optic nerve development(GO:0021554) |
| 0.0 |
0.0 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 |
0.1 |
GO:0045948 |
positive regulation of translational initiation(GO:0045948) |
| 0.0 |
0.2 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.0 |
0.1 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.0 |
0.1 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.0 |
0.1 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.0 |
0.3 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.0 |
0.2 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
0.0 |
GO:1990776 |
angiotensin-activated signaling pathway(GO:0038166) cellular response to angiotensin(GO:1904385) response to angiotensin(GO:1990776) |
| 0.0 |
0.1 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.1 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.0 |
GO:0031053 |
adenosine to inosine editing(GO:0006382) primary miRNA processing(GO:0031053) |
| 0.0 |
0.1 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.0 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 |
0.3 |
GO:0001895 |
retina homeostasis(GO:0001895) |
| 0.0 |
0.2 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.0 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 |
0.1 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.3 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.1 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 |
0.1 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.3 |
GO:2000648 |
positive regulation of stem cell proliferation(GO:2000648) |