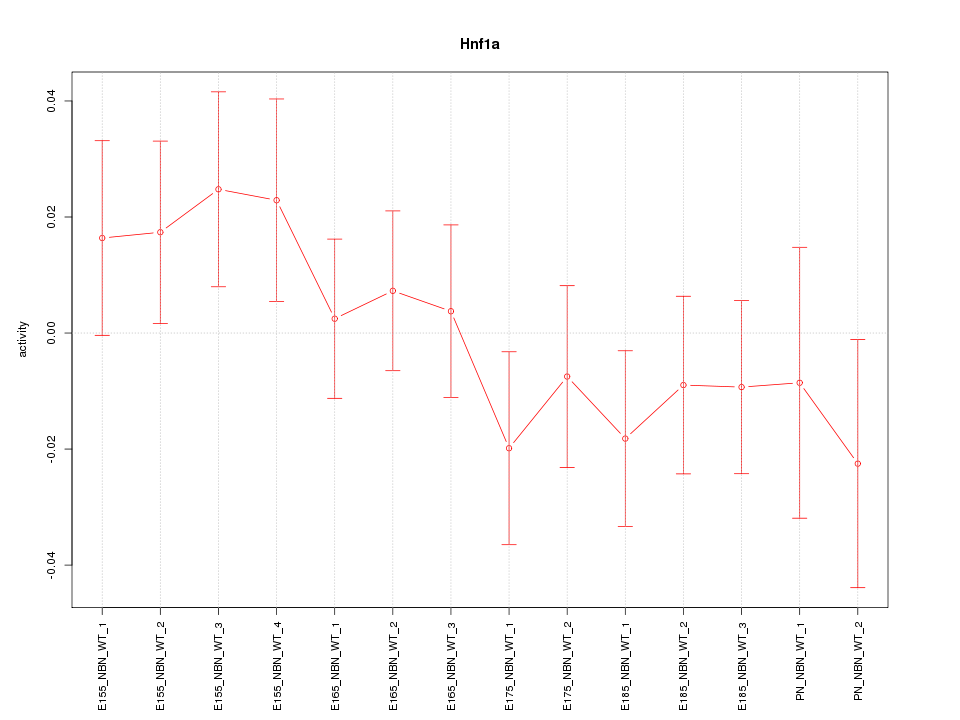
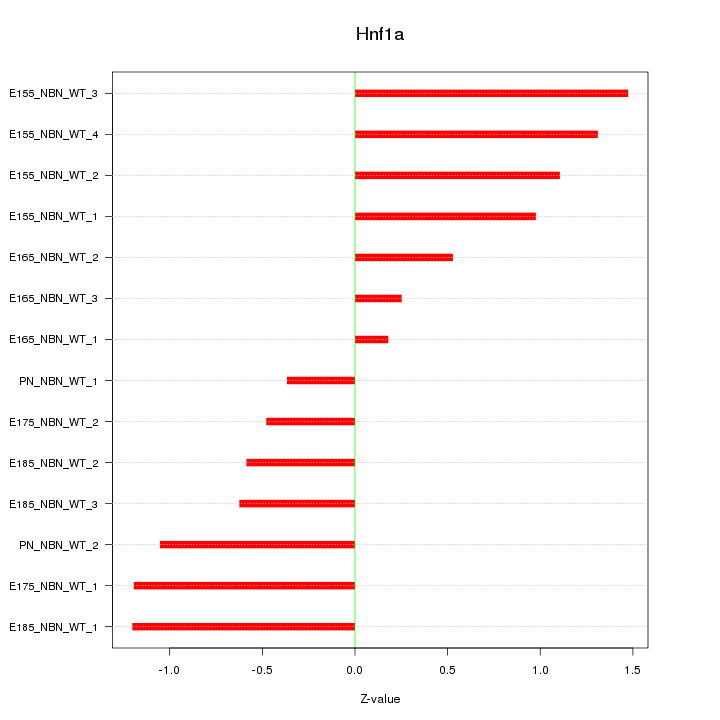
Motif ID: Hnf1a
Z-value: 0.907
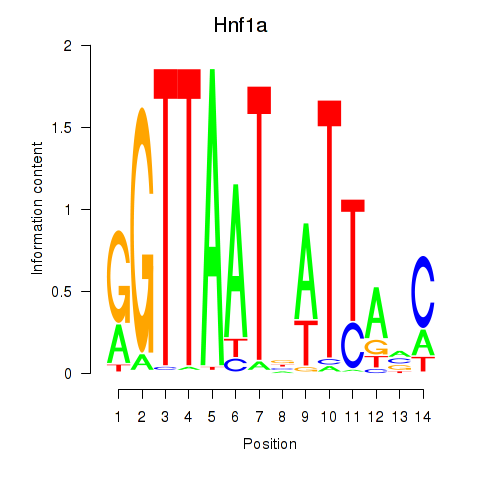
Transcription factors associated with Hnf1a:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hnf1a | ENSMUSG00000029556.6 | Hnf1a |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.8 | 3.3 | GO:0032275 | luteinizing hormone secretion(GO:0032275) positive regulation of gonadotropin secretion(GO:0032278) |
| 0.6 | 1.8 | GO:1901731 | calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.4 | 1.2 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.4 | 1.9 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.4 | 1.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 | 2.0 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.3 | 0.8 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.2 | 0.9 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.1 | 0.8 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.8 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 1.0 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.3 | GO:0032782 | urea transport(GO:0015840) bile acid secretion(GO:0032782) urea transmembrane transport(GO:0071918) |
| 0.0 | 1.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.3 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 14.4 | GO:0016055 | Wnt signaling pathway(GO:0016055) cell-cell signaling by wnt(GO:0198738) |
| 0.0 | 0.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 2.9 | GO:0042384 | cilium assembly(GO:0042384) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.3 | 0.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 0.9 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 2.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 4.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 3.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 17.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.2 | 3.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 2.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 1.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.9 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |