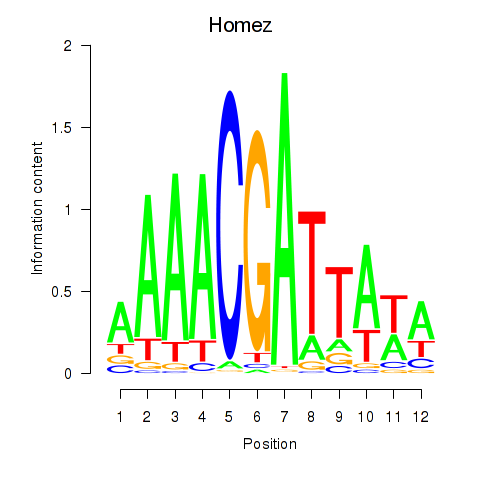
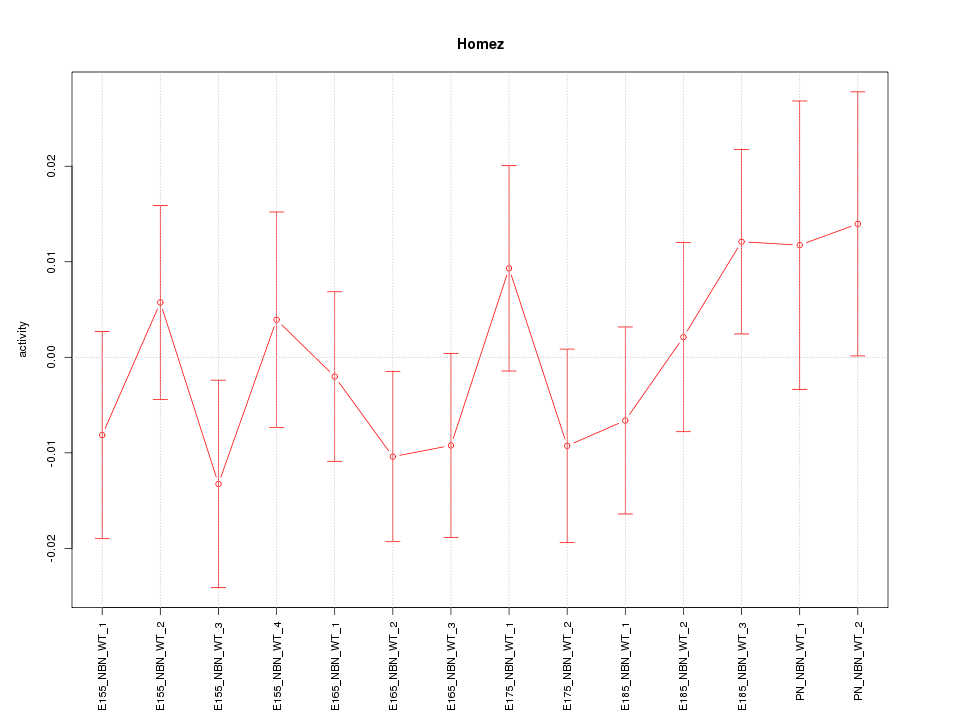
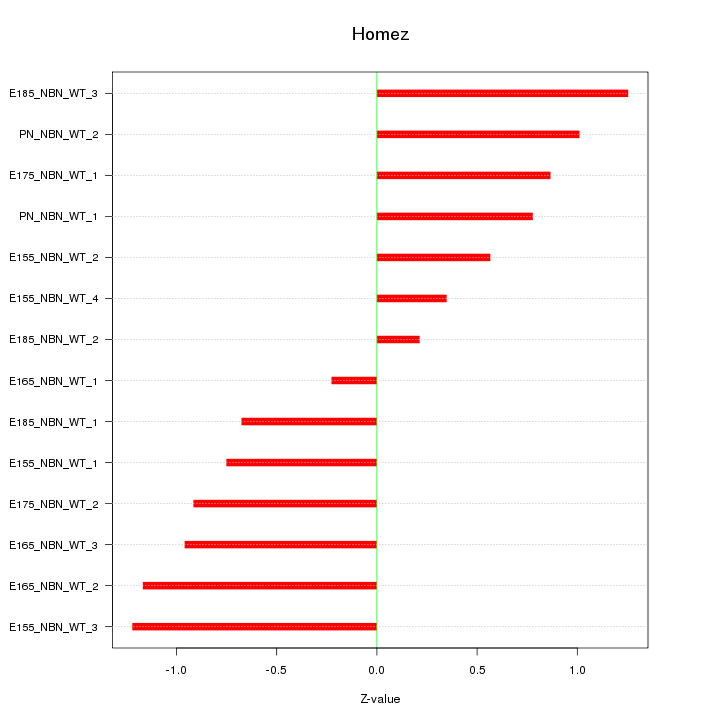
| 0.4 |
1.7 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 0.4 |
1.2 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.3 |
1.2 |
GO:1902608 |
regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.3 |
0.8 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.3 |
0.8 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.2 |
0.5 |
GO:0060217 |
hemangioblast cell differentiation(GO:0060217) |
| 0.2 |
0.8 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 |
0.5 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.1 |
0.4 |
GO:0040009 |
regulation of growth rate(GO:0040009) |
| 0.1 |
0.3 |
GO:0050713 |
negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 |
0.7 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 |
0.6 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.3 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.1 |
0.3 |
GO:0002765 |
immune response-inhibiting signal transduction(GO:0002765) |
| 0.1 |
0.6 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.1 |
0.7 |
GO:1901387 |
positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 |
0.7 |
GO:0010793 |
regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.1 |
0.6 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 |
0.3 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.1 |
0.7 |
GO:0031999 |
negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 |
0.2 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 |
0.3 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 |
0.5 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 |
0.2 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 |
0.2 |
GO:0016321 |
female meiosis chromosome segregation(GO:0016321) |
| 0.1 |
0.3 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.1 |
0.2 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 |
0.2 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 |
0.2 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 |
0.3 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.3 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 |
0.2 |
GO:0002606 |
positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 |
0.3 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.4 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.7 |
GO:0060628 |
regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 |
1.2 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.3 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 |
0.4 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 |
0.3 |
GO:0035999 |
tetrahydrofolate interconversion(GO:0035999) |
| 0.0 |
0.1 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 |
0.5 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 |
0.2 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 |
0.1 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.0 |
0.3 |
GO:0035280 |
miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 |
0.1 |
GO:0045448 |
regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 |
0.9 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 0.0 |
0.2 |
GO:0010745 |
negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 |
2.7 |
GO:0048663 |
neuron fate commitment(GO:0048663) |
| 0.0 |
1.1 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.3 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 |
0.3 |
GO:0032367 |
intracellular cholesterol transport(GO:0032367) |
| 0.0 |
0.7 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.2 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.1 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.0 |
0.6 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 |
0.7 |
GO:0033962 |
cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 |
0.1 |
GO:0045065 |
cytotoxic T cell differentiation(GO:0045065) |
| 0.0 |
0.1 |
GO:0034241 |
positive regulation of macrophage fusion(GO:0034241) regulation of osteoclast proliferation(GO:0090289) |
| 0.0 |
0.1 |
GO:0046121 |
deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 |
0.1 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.0 |
0.0 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 |
0.7 |
GO:0046825 |
regulation of protein export from nucleus(GO:0046825) |
| 0.0 |
0.4 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.0 |
0.2 |
GO:0006366 |
transcription from RNA polymerase II promoter(GO:0006366) |
| 0.0 |
0.1 |
GO:0070836 |
caveola assembly(GO:0070836) |
| 0.0 |
1.0 |
GO:0060291 |
long-term synaptic potentiation(GO:0060291) |
| 0.0 |
0.4 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
0.2 |
GO:0009651 |
response to salt stress(GO:0009651) |
| 0.0 |
0.1 |
GO:1904321 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 |
0.1 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 |
0.2 |
GO:0048015 |
phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.0 |
0.7 |
GO:0008543 |
fibroblast growth factor receptor signaling pathway(GO:0008543) |