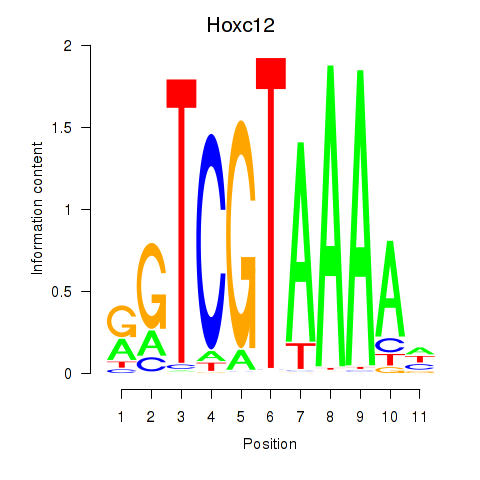
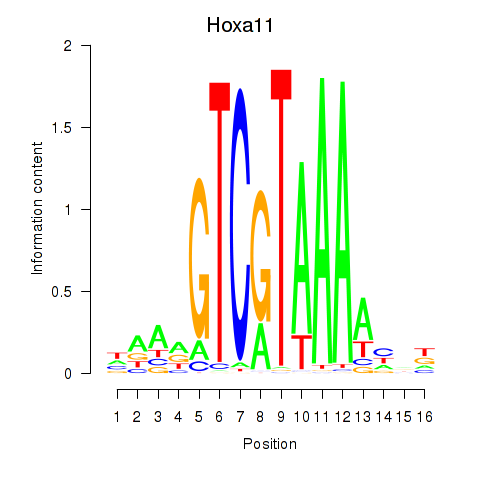
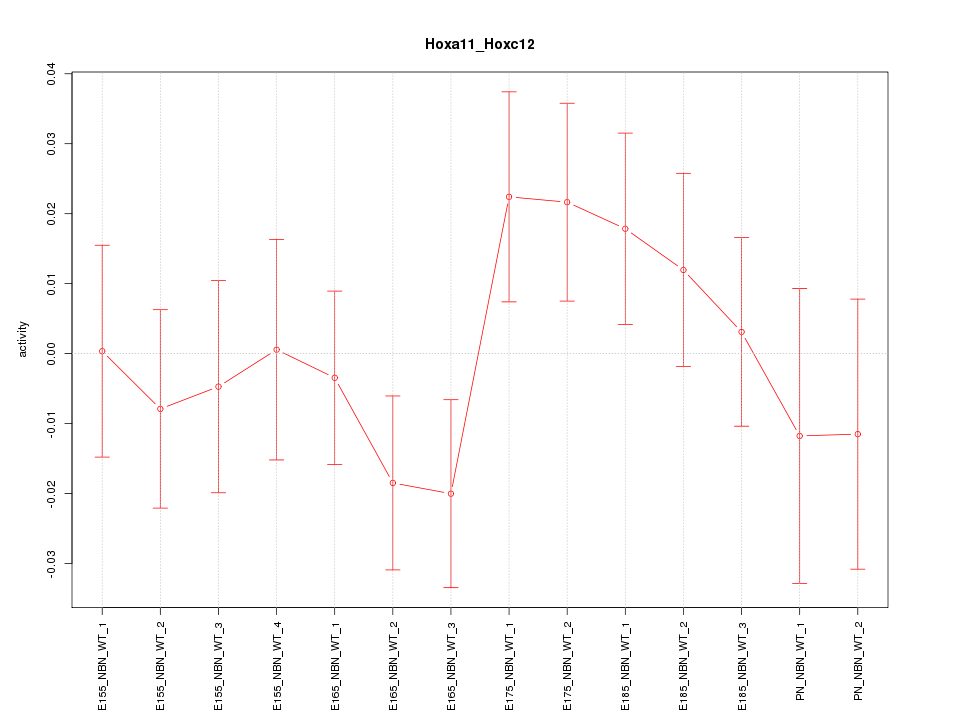
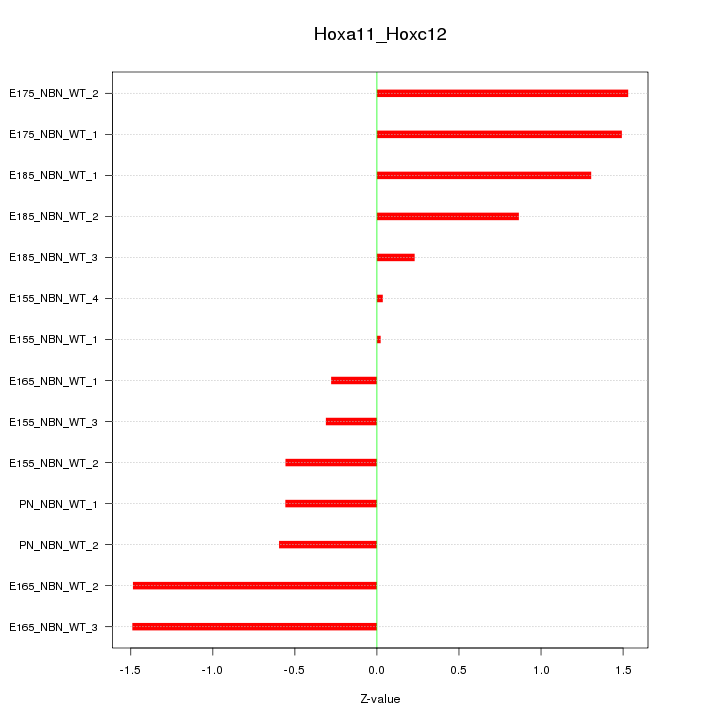
| 1.4 |
4.2 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.3 |
1.0 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.2 |
0.9 |
GO:0072362 |
regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.2 |
1.2 |
GO:0042045 |
epithelial fluid transport(GO:0042045) |
| 0.2 |
0.6 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.2 |
0.6 |
GO:0044861 |
protein transport into plasma membrane raft(GO:0044861) |
| 0.1 |
0.6 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.1 |
0.3 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 |
1.1 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.5 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.7 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.1 |
0.8 |
GO:0031999 |
negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 |
0.8 |
GO:0036120 |
cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 |
0.4 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.1 |
0.2 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 |
0.2 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 |
0.2 |
GO:0048352 |
neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.1 |
0.2 |
GO:0043201 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 |
0.2 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.5 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.4 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.0 |
0.3 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.0 |
0.1 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.0 |
0.1 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.0 |
0.5 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 |
0.2 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.0 |
0.4 |
GO:0043030 |
regulation of macrophage activation(GO:0043030) |
| 0.0 |
0.1 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.2 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.1 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 |
0.2 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.0 |
0.1 |
GO:0051771 |
negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 |
0.3 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.2 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.0 |
0.2 |
GO:0033523 |
endodermal cell fate commitment(GO:0001711) histone H2B ubiquitination(GO:0033523) |