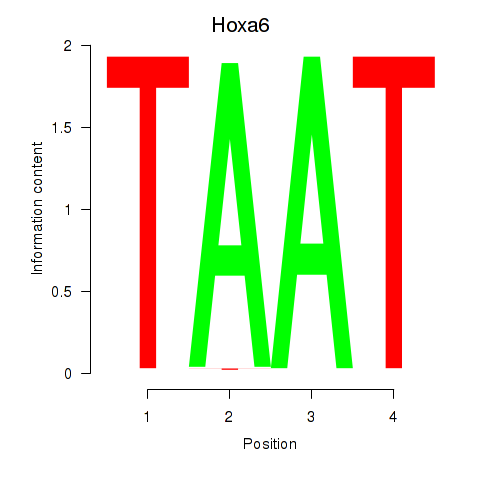
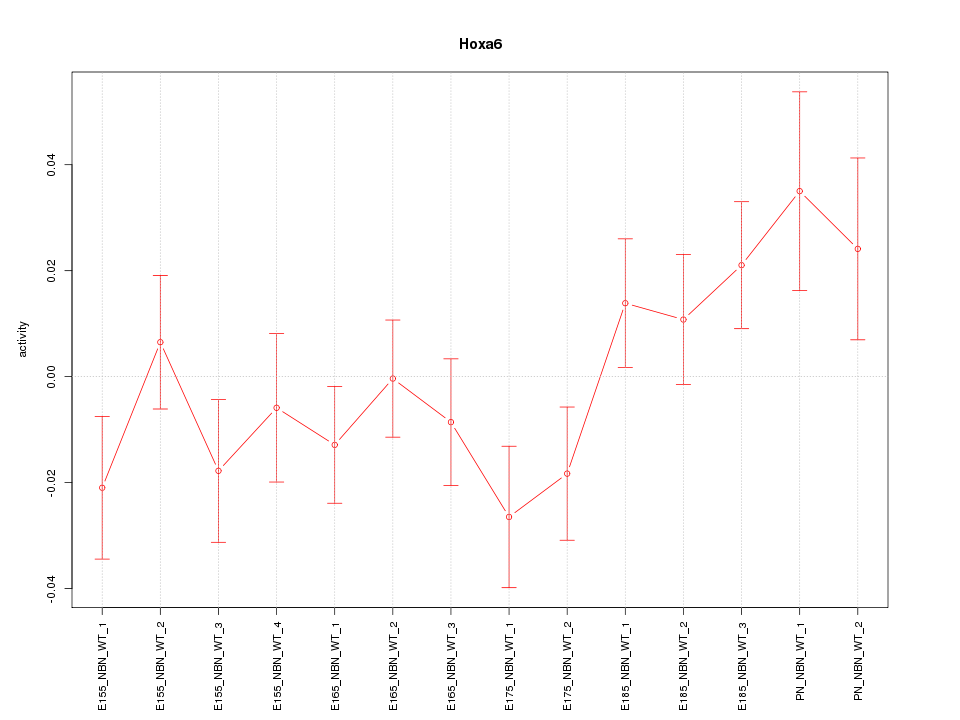
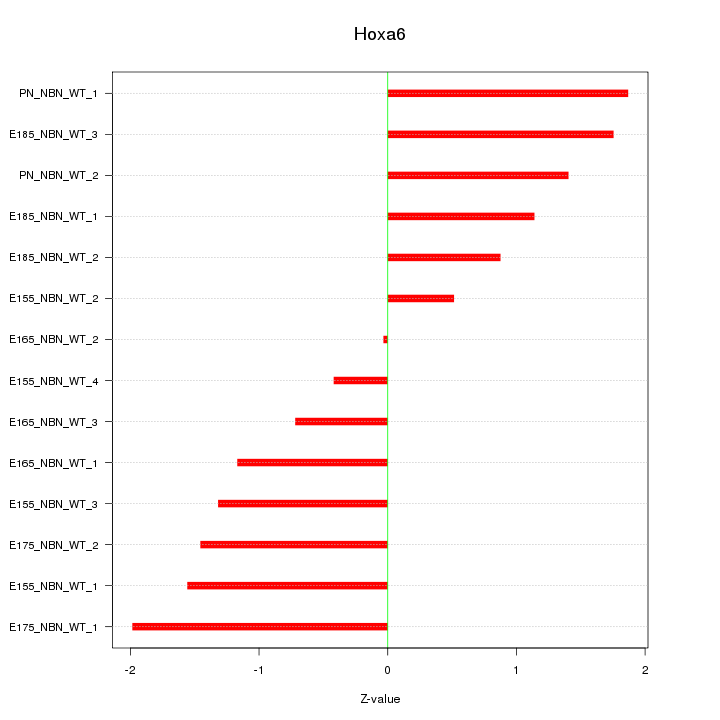
| 4.4 |
13.2 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.8 |
7.0 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.1 |
3.3 |
GO:0060023 |
soft palate development(GO:0060023) |
| 1.0 |
3.9 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.9 |
4.5 |
GO:0061047 |
positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.9 |
6.1 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.7 |
2.0 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.5 |
3.5 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.5 |
1.9 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.5 |
3.2 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.4 |
2.4 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.4 |
3.2 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.4 |
1.2 |
GO:1990314 |
cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.4 |
14.0 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.4 |
1.5 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 |
1.6 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 |
0.7 |
GO:2001015 |
G1 to G0 transition involved in cell differentiation(GO:0070315) negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.2 |
1.2 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.2 |
2.4 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 |
0.7 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 |
1.8 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 |
0.6 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 0.2 |
1.2 |
GO:0032229 |
positive regulation of gamma-aminobutyric acid secretion(GO:0014054) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.2 |
1.3 |
GO:1900747 |
negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 |
6.1 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.2 |
0.4 |
GO:0090245 |
axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 |
4.3 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.2 |
2.1 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 |
0.3 |
GO:0021562 |
vestibulocochlear nerve development(GO:0021562) |
| 0.2 |
1.2 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 |
1.0 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.2 |
1.0 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.2 |
3.8 |
GO:0036342 |
post-anal tail morphogenesis(GO:0036342) |
| 0.2 |
5.9 |
GO:0010259 |
multicellular organism aging(GO:0010259) |
| 0.2 |
6.4 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.1 |
2.3 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 |
1.3 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.1 |
3.2 |
GO:0042474 |
middle ear morphogenesis(GO:0042474) |
| 0.1 |
1.5 |
GO:0021800 |
cerebral cortex tangential migration(GO:0021800) |
| 0.1 |
3.2 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.1 |
0.7 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.1 |
0.3 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.1 |
0.5 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.1 |
0.2 |
GO:0061303 |
oculomotor nerve development(GO:0021557) cornea development in camera-type eye(GO:0061303) |
| 0.1 |
0.8 |
GO:0002118 |
aggressive behavior(GO:0002118) |
| 0.1 |
4.7 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.1 |
1.8 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.1 |
0.3 |
GO:0010835 |
regulation of protein ADP-ribosylation(GO:0010835) |
| 0.1 |
0.6 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.8 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.1 |
1.4 |
GO:0048665 |
neuron fate specification(GO:0048665) |
| 0.1 |
1.0 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.1 |
0.3 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 |
0.3 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.7 |
GO:0009048 |
dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 |
0.2 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 |
1.2 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
1.3 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.1 |
GO:0003150 |
muscular septum morphogenesis(GO:0003150) |
| 0.0 |
2.2 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.9 |
GO:0006625 |
protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 |
0.2 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
2.0 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.6 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.3 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
2.6 |
GO:0007605 |
sensory perception of sound(GO:0007605) |
| 0.0 |
0.3 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.8 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.8 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 |
0.2 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.4 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 |
0.4 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 |
1.0 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
0.3 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.0 |
1.8 |
GO:0021987 |
cerebral cortex development(GO:0021987) |
| 0.0 |
0.8 |
GO:0007566 |
embryo implantation(GO:0007566) |
| 0.0 |
0.3 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.6 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.2 |
GO:0031987 |
locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 |
0.4 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |