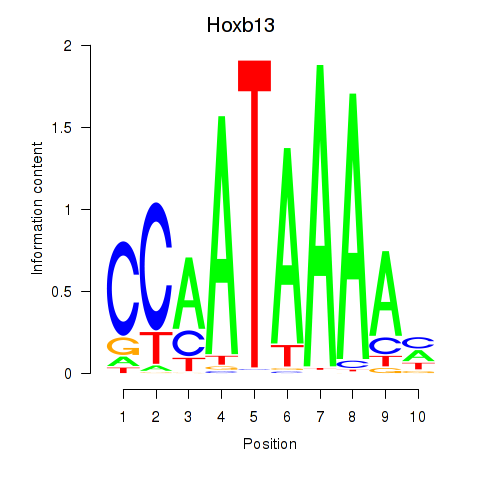
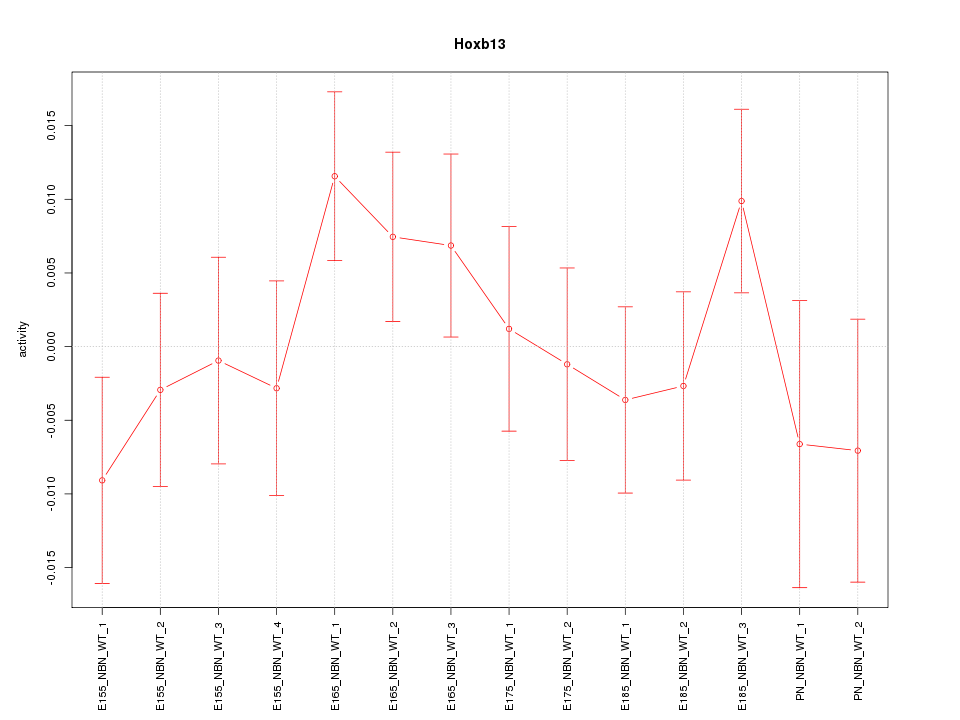
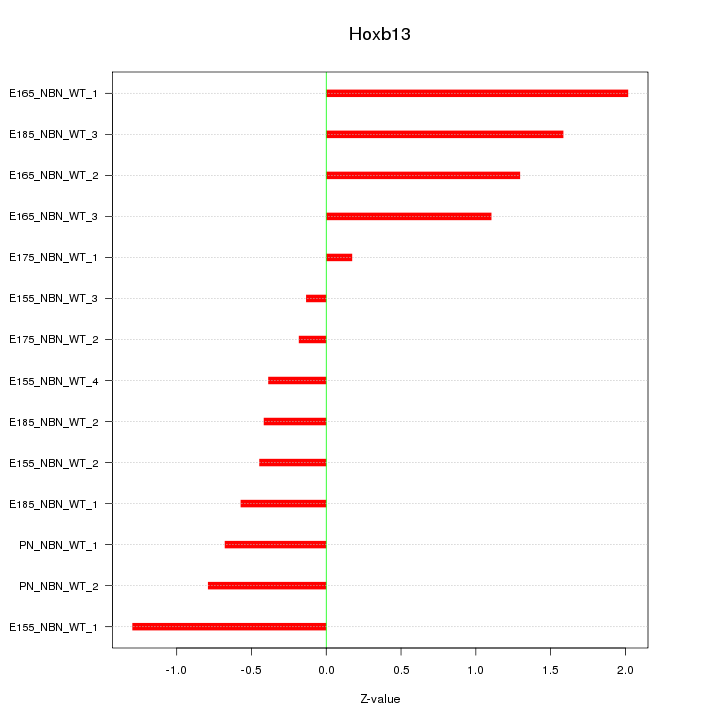
| 0.2 |
0.7 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.2 |
0.6 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.2 |
0.6 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.2 |
0.2 |
GO:0097094 |
craniofacial suture morphogenesis(GO:0097094) |
| 0.2 |
0.6 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.2 |
0.5 |
GO:0035604 |
fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.2 |
0.7 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.2 |
0.5 |
GO:0048698 |
negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.2 |
0.5 |
GO:0071873 |
response to norepinephrine(GO:0071873) |
| 0.2 |
0.5 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.2 |
0.5 |
GO:2000409 |
positive regulation of T cell extravasation(GO:2000409) |
| 0.1 |
0.6 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 |
0.4 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 |
0.6 |
GO:1900533 |
medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 |
0.5 |
GO:1901204 |
positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.1 |
0.4 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 |
0.5 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 |
0.4 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.6 |
GO:0002121 |
inter-male aggressive behavior(GO:0002121) |
| 0.1 |
0.1 |
GO:0045763 |
negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 |
0.3 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 |
0.5 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.1 |
0.3 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) |
| 0.1 |
0.4 |
GO:0055095 |
lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 |
0.4 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.1 |
0.3 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.1 |
0.1 |
GO:0023041 |
neuronal signal transduction(GO:0023041) |
| 0.1 |
0.3 |
GO:0060523 |
prostate epithelial cord elongation(GO:0060523) |
| 0.1 |
0.5 |
GO:1902956 |
regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.1 |
1.1 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.3 |
GO:0006166 |
purine ribonucleoside salvage(GO:0006166) |
| 0.1 |
0.2 |
GO:0010636 |
positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 |
0.3 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 |
0.2 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.1 |
1.2 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 |
0.2 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 |
0.3 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 |
0.2 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.1 |
0.5 |
GO:0061502 |
early endosome to recycling endosome transport(GO:0061502) |
| 0.1 |
0.2 |
GO:0070649 |
formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 |
0.2 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 |
0.2 |
GO:0016256 |
N-glycan processing to lysosome(GO:0016256) |
| 0.1 |
0.3 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 |
0.2 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 |
0.1 |
GO:2000525 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 |
0.1 |
GO:0046502 |
uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 |
0.3 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 |
0.3 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
0.4 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.1 |
0.6 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.4 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 |
0.5 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.1 |
0.2 |
GO:0070086 |
ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 |
0.3 |
GO:1901164 |
negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 |
0.5 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.1 |
0.3 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.1 |
0.2 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.1 |
0.3 |
GO:1902856 |
negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 |
0.2 |
GO:0070428 |
granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 |
0.2 |
GO:0032382 |
positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.1 |
0.1 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.1 |
0.6 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.1 |
0.3 |
GO:2000767 |
positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 |
0.3 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.4 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 |
0.5 |
GO:0000972 |
transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 |
0.2 |
GO:0060282 |
positive regulation of oocyte development(GO:0060282) |
| 0.1 |
0.2 |
GO:0009838 |
abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 |
0.1 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) |
| 0.0 |
0.4 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.2 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.0 |
0.7 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.0 |
0.2 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
0.3 |
GO:0038180 |
nerve growth factor signaling pathway(GO:0038180) |
| 0.0 |
0.2 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.0 |
0.1 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.0 |
0.2 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.0 |
0.5 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.2 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 |
0.2 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 |
0.2 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 |
0.4 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
1.2 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.7 |
GO:0060712 |
spongiotrophoblast layer development(GO:0060712) |
| 0.0 |
0.1 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 |
0.2 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.2 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.0 |
0.3 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.0 |
0.8 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.2 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.0 |
0.2 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.1 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) |
| 0.0 |
0.4 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 |
0.3 |
GO:0009162 |
nucleoside monophosphate catabolic process(GO:0009125) deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 |
0.2 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.0 |
0.2 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.0 |
0.2 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.0 |
0.3 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 |
0.3 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 |
0.2 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.0 |
0.3 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.0 |
0.1 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.0 |
0.1 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 |
0.6 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.3 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.1 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 |
0.2 |
GO:0006826 |
iron ion transport(GO:0006826) |
| 0.0 |
0.1 |
GO:0038108 |
negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 |
0.1 |
GO:0035552 |
oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 |
0.2 |
GO:1901727 |
positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 |
0.2 |
GO:0015867 |
ATP transport(GO:0015867) |
| 0.0 |
0.4 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.3 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.3 |
GO:1902414 |
protein localization to cell junction(GO:1902414) |
| 0.0 |
0.1 |
GO:0008594 |
photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 |
0.1 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 |
0.4 |
GO:0061162 |
establishment of monopolar cell polarity(GO:0061162) |
| 0.0 |
0.3 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.3 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.2 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 |
0.1 |
GO:1902445 |
regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 |
0.3 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.5 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.2 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 |
0.1 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 |
0.1 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.0 |
0.1 |
GO:0031038 |
myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.0 |
0.1 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 |
0.2 |
GO:0060315 |
negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 |
0.3 |
GO:0009396 |
folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 |
0.3 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.3 |
GO:0070234 |
positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 |
0.6 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 |
0.3 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 |
0.1 |
GO:0070375 |
ERK5 cascade(GO:0070375) |
| 0.0 |
0.8 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.0 |
0.1 |
GO:0045410 |
positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 |
0.1 |
GO:0042776 |
mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 |
0.1 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.3 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.1 |
GO:0001193 |
maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.0 |
0.1 |
GO:1990034 |
calcium ion export from cell(GO:1990034) calcium ion import into cell(GO:1990035) |
| 0.0 |
0.2 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.1 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 |
0.1 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.2 |
GO:0045040 |
protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 |
0.3 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 0.0 |
0.1 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 |
0.1 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 |
0.1 |
GO:0071864 |
positive regulation of cell proliferation in bone marrow(GO:0071864) |
| 0.0 |
0.3 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 |
0.2 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.0 |
0.6 |
GO:1902259 |
regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 |
0.1 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 |
0.2 |
GO:0060371 |
regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 |
0.2 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.0 |
0.4 |
GO:0014829 |
vascular smooth muscle contraction(GO:0014829) |
| 0.0 |
0.1 |
GO:2001268 |
negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 |
0.2 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.1 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.0 |
0.1 |
GO:0036089 |
cleavage furrow formation(GO:0036089) |
| 0.0 |
0.5 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.0 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.0 |
0.1 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 |
0.2 |
GO:0030174 |
regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 |
0.2 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
1.7 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.0 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 |
0.1 |
GO:0046133 |
pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.0 |
0.1 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.0 |
0.1 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 |
0.2 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 |
0.1 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 |
0.1 |
GO:0097475 |
motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 |
0.1 |
GO:0019441 |
tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) quinolinate biosynthetic process(GO:0019805) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.0 |
0.1 |
GO:0051775 |
response to redox state(GO:0051775) |
| 0.0 |
0.1 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 |
0.2 |
GO:0090308 |
regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 |
0.3 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.1 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.0 |
0.2 |
GO:0060828 |
regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 |
0.3 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
0.1 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.0 |
0.4 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.0 |
0.3 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:0070126 |
mitochondrial translational termination(GO:0070126) |
| 0.0 |
0.1 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.0 |
0.0 |
GO:0042726 |
flavin-containing compound metabolic process(GO:0042726) |
| 0.0 |
0.1 |
GO:0035902 |
response to immobilization stress(GO:0035902) |
| 0.0 |
1.1 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.3 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.0 |
0.1 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 |
0.1 |
GO:0006537 |
glutamate biosynthetic process(GO:0006537) |
| 0.0 |
0.0 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.0 |
0.3 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.0 |
GO:1902512 |
positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 |
0.2 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.1 |
GO:0042756 |
drinking behavior(GO:0042756) |
| 0.0 |
0.1 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 |
0.1 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.0 |
0.1 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.1 |
GO:1902177 |
positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 |
0.9 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.6 |
GO:1901381 |
positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.1 |
GO:1900748 |
positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 |
0.1 |
GO:2000074 |
regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 |
0.1 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.0 |
0.1 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.2 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.1 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 |
0.1 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.1 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.0 |
0.3 |
GO:0032094 |
response to activity(GO:0014823) response to food(GO:0032094) |
| 0.0 |
0.3 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 |
0.5 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.8 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.1 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 |
0.1 |
GO:0090315 |
negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 |
0.5 |
GO:1902230 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 |
0.2 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.1 |
GO:0007320 |
insemination(GO:0007320) |
| 0.0 |
0.0 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.0 |
0.2 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.0 |
GO:0036518 |
chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 |
0.1 |
GO:0003009 |
skeletal muscle contraction(GO:0003009) |
| 0.0 |
0.2 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.0 |
0.1 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 |
0.1 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.1 |
GO:0006824 |
cobalt ion transport(GO:0006824) |
| 0.0 |
0.1 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 |
0.1 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.2 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.2 |
GO:0045648 |
positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 |
0.1 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.3 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.1 |
GO:0018158 |
protein oxidation(GO:0018158) |
| 0.0 |
0.3 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.1 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 |
0.1 |
GO:0015862 |
uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 |
0.1 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.0 |
0.0 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 |
0.1 |
GO:0051573 |
negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 |
0.2 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.2 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.1 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 |
0.3 |
GO:0014742 |
positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 |
1.1 |
GO:0050821 |
protein stabilization(GO:0050821) |
| 0.0 |
0.1 |
GO:0070498 |
interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 |
0.1 |
GO:0000467 |
exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 |
0.1 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.1 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 |
0.1 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.1 |
GO:0008211 |
glucocorticoid metabolic process(GO:0008211) |
| 0.0 |
0.0 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.0 |
0.1 |
GO:0007602 |
phototransduction(GO:0007602) |