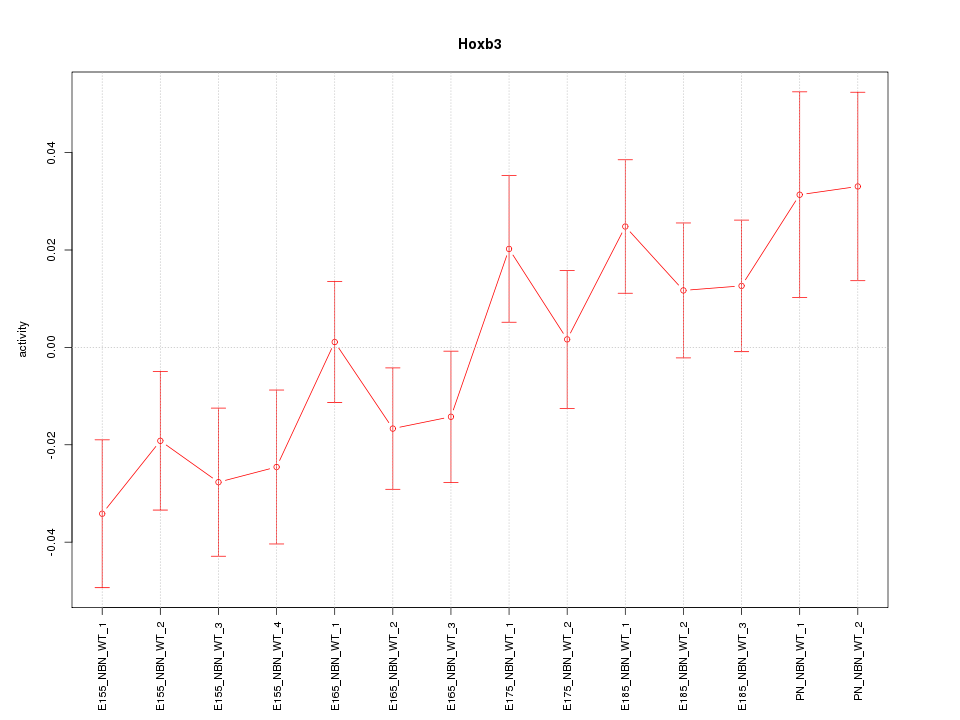
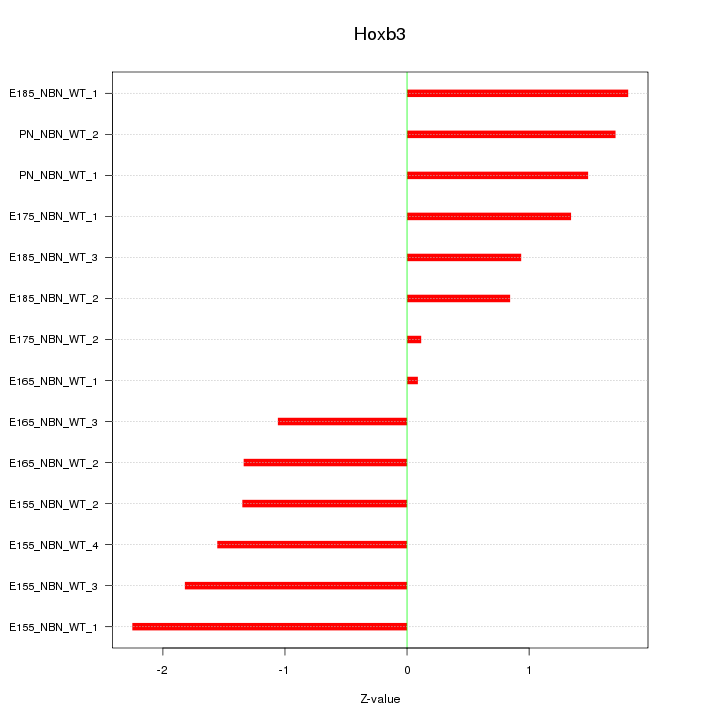
Motif ID: Hoxb3
Z-value: 1.397
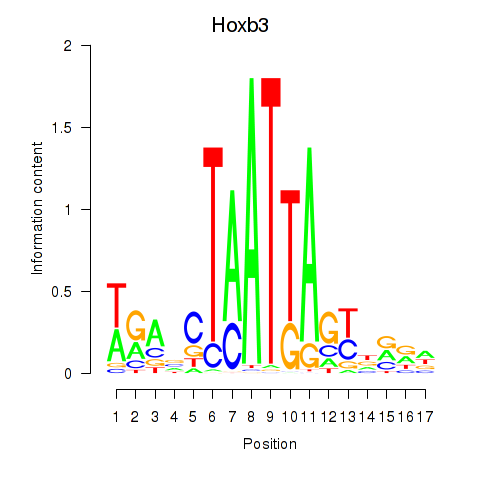
Transcription factors associated with Hoxb3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxb3 | ENSMUSG00000048763.5 | Hoxb3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0030091 | protein repair(GO:0030091) |
| 1.0 | 3.9 | GO:1900272 | negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.8 | 2.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.7 | 2.9 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.6 | 1.9 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.5 | 3.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.4 | 1.5 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.4 | 2.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 1.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.3 | 1.9 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 0.8 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 1.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 3.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 3.9 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 2.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 6.6 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.2 | 0.9 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.2 | 0.6 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 0.9 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 0.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 3.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.4 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 | 0.4 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.1 | 0.8 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 1.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.0 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 2.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.3 | GO:0090403 | T cell proliferation involved in immune response(GO:0002309) regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) regulation of fermentation(GO:0043465) oxidative stress-induced premature senescence(GO:0090403) negative regulation of fermentation(GO:1901003) |
| 0.1 | 1.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 6.1 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 0.4 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.2 | GO:2000338 | positive regulation of interleukin-23 production(GO:0032747) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.1 | 0.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 1.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.9 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.6 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 2.6 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 3.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.5 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 7.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 3.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 2.9 | GO:0043205 | fibril(GO:0043205) |
| 0.2 | 1.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 3.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 2.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 8.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 1.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 2.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.6 | 2.9 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.5 | 5.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.5 | 1.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.5 | 2.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.5 | 5.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 3.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.4 | 1.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.4 | 1.5 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.3 | 0.8 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.2 | 1.9 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 2.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 1.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 3.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 1.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 3.0 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 1.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.0 | 1.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 3.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 3.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 2.8 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:0001221 | transcription cofactor binding(GO:0001221) |