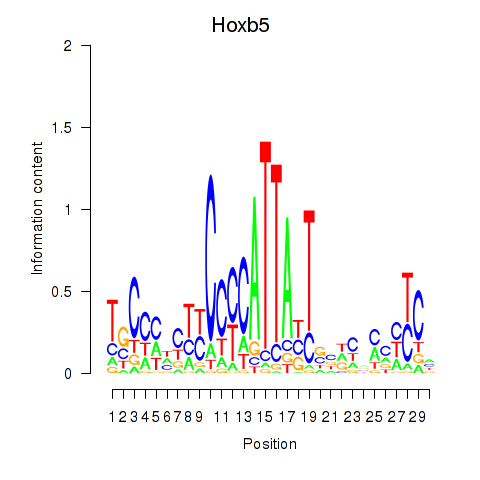
| 0.3 |
1.2 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.2 |
0.8 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.2 |
1.2 |
GO:0071361 |
cellular response to zinc ion(GO:0071294) cellular response to ethanol(GO:0071361) |
| 0.1 |
0.4 |
GO:0070649 |
formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 |
0.4 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.1 |
2.1 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
0.3 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.1 |
0.5 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 |
0.3 |
GO:0010248 |
B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 |
1.7 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.1 |
1.6 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.1 |
0.3 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 |
0.3 |
GO:2000823 |
regulation of androgen receptor activity(GO:2000823) |
| 0.1 |
0.2 |
GO:0070172 |
positive regulation of tooth mineralization(GO:0070172) |
| 0.1 |
0.2 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.0 |
0.2 |
GO:0097374 |
sensory neuron axon guidance(GO:0097374) |
| 0.0 |
0.4 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 |
0.4 |
GO:0000963 |
mitochondrial RNA processing(GO:0000963) |
| 0.0 |
0.4 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.2 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.9 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.8 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 |
1.0 |
GO:0003016 |
respiratory system process(GO:0003016) |
| 0.0 |
0.2 |
GO:0045583 |
regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 |
1.3 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.0 |
0.3 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
0.7 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.2 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.6 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.4 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.2 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.0 |
0.4 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.3 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.7 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
0.1 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |