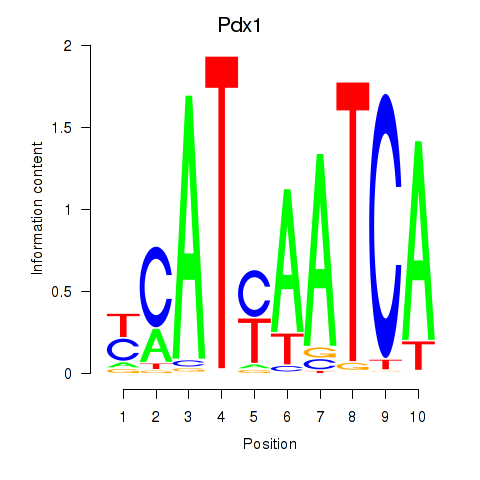
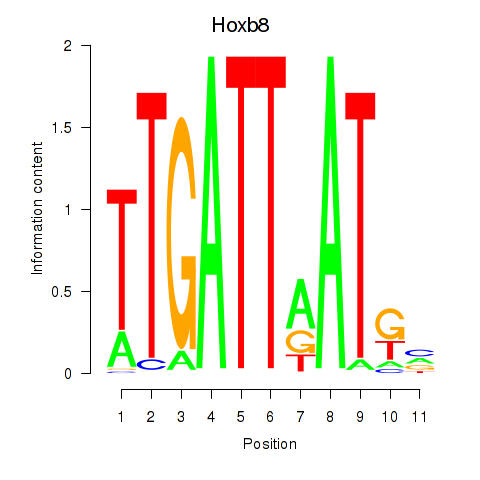
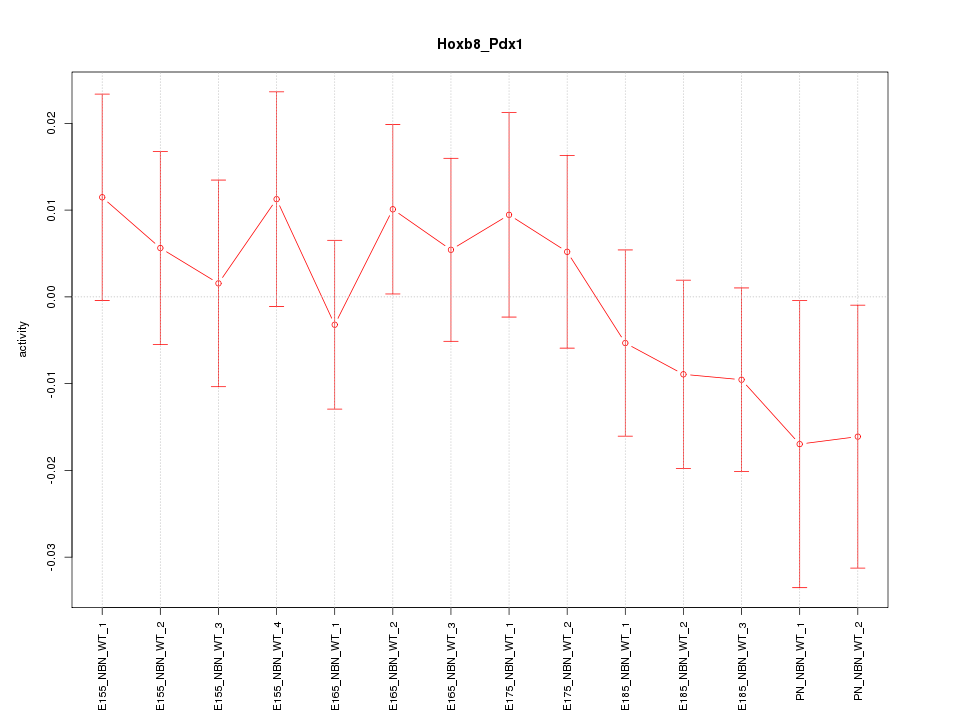
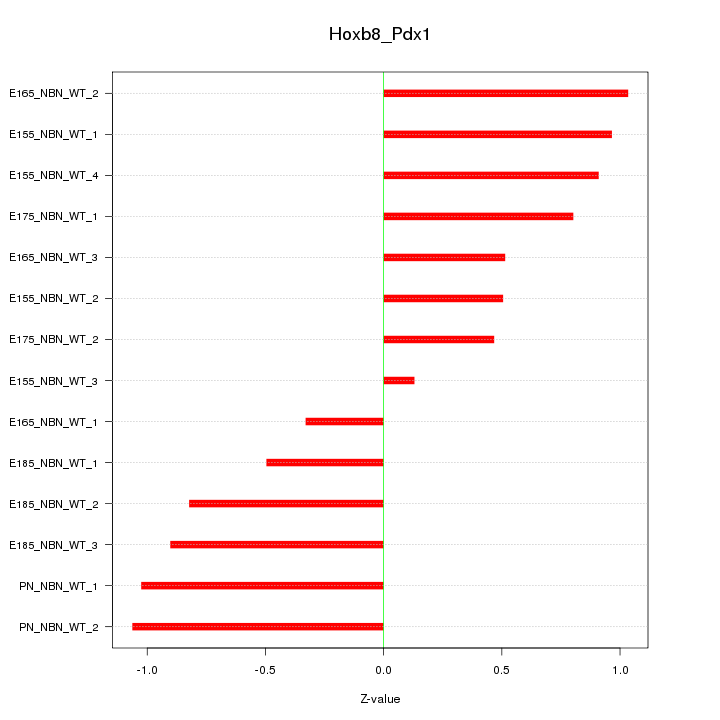
| 0.9 |
2.6 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.5 |
1.8 |
GO:0055095 |
lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.4 |
3.4 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.4 |
5.4 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 |
1.0 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 |
0.7 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.2 |
0.7 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.2 |
0.9 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.2 |
2.1 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.2 |
3.4 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.2 |
1.7 |
GO:0060373 |
regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 |
2.4 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 |
2.4 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.2 |
0.3 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 |
0.7 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 |
0.8 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 |
0.8 |
GO:0046549 |
retinal cone cell development(GO:0046549) |
| 0.1 |
1.0 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 |
0.6 |
GO:1900242 |
regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 |
0.9 |
GO:0048852 |
diencephalon morphogenesis(GO:0048852) |
| 0.1 |
1.0 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 |
1.7 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 |
1.3 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 |
0.5 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.1 |
0.4 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 |
1.8 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 |
0.6 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.1 |
0.6 |
GO:0034720 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) histone H3-K4 demethylation(GO:0034720) mammary duct terminal end bud growth(GO:0060763) |
| 0.1 |
0.2 |
GO:0042264 |
peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 |
1.6 |
GO:0021692 |
cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.1 |
0.5 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 |
0.2 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.1 |
0.2 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.0 |
0.4 |
GO:0060484 |
lung-associated mesenchyme development(GO:0060484) |
| 0.0 |
0.1 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.0 |
1.1 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.3 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.3 |
GO:0032693 |
negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 |
0.6 |
GO:0032506 |
cytokinetic process(GO:0032506) |
| 0.0 |
0.2 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 |
3.1 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.1 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 |
0.2 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.2 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.0 |
0.7 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 |
0.3 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 |
0.5 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.0 |
0.2 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.5 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 |
0.4 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) |
| 0.0 |
0.6 |
GO:0045725 |
positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 |
0.1 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 |
0.2 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.0 |
0.2 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 |
1.4 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.2 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.4 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.6 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.1 |
GO:0043987 |
histone-serine phosphorylation(GO:0035404) histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 |
0.2 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.2 |
GO:0017121 |
phospholipid scrambling(GO:0017121) |
| 0.0 |
0.1 |
GO:0051030 |
snRNA transport(GO:0051030) |
| 0.0 |
0.3 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.0 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.1 |
GO:0031622 |
positive regulation of fever generation(GO:0031622) negative regulation of action potential(GO:0045759) |
| 0.0 |
0.1 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.0 |
0.2 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
0.2 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
1.3 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.1 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 |
0.5 |
GO:0043123 |
positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 |
0.2 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 |
0.3 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.0 |
0.2 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.0 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 |
0.2 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.6 |
GO:0042168 |
heme metabolic process(GO:0042168) |
| 0.0 |
0.2 |
GO:0010715 |
regulation of extracellular matrix disassembly(GO:0010715) |
| 0.0 |
1.0 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.0 |
GO:0044778 |
meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 |
0.2 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.6 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.3 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.2 |
GO:0090140 |
regulation of mitochondrial fission(GO:0090140) |
| 0.0 |
0.0 |
GO:0046689 |
response to mercury ion(GO:0046689) |