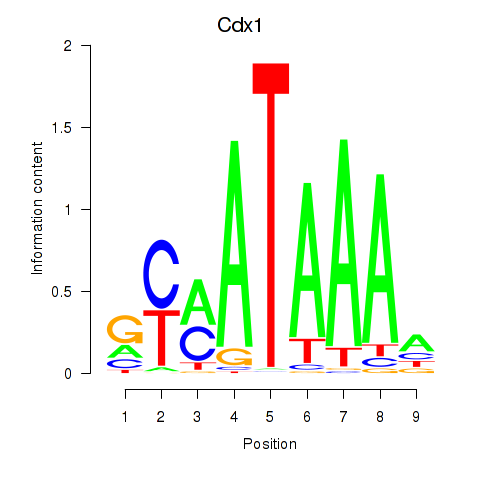
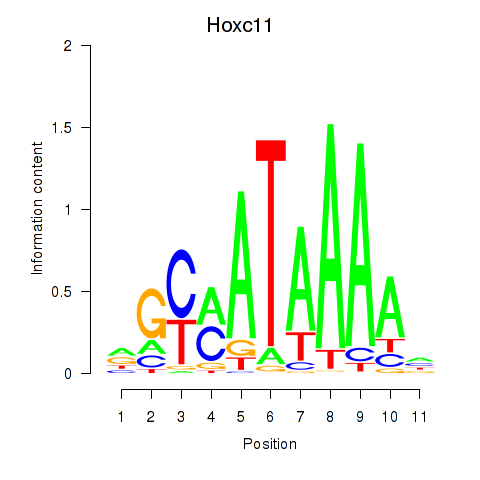
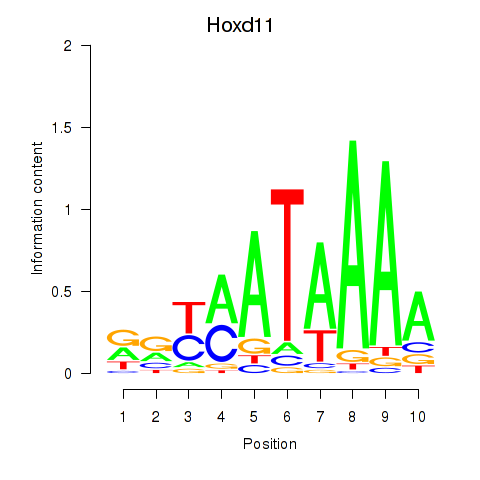
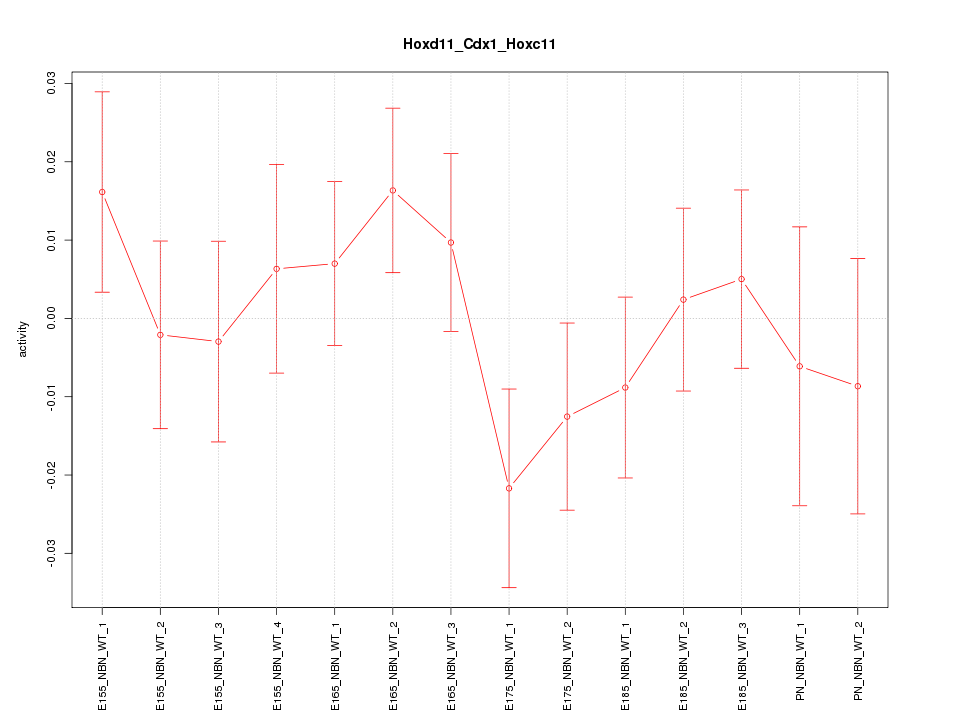
| 2.0 |
16.2 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.7 |
2.0 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 0.4 |
1.1 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.3 |
1.5 |
GO:0060467 |
negative regulation of fertilization(GO:0060467) |
| 0.3 |
1.2 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.2 |
1.6 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.2 |
0.7 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.2 |
0.6 |
GO:0046098 |
regulation of primitive erythrocyte differentiation(GO:0010725) guanine metabolic process(GO:0046098) |
| 0.2 |
0.6 |
GO:0006526 |
arginine biosynthetic process(GO:0006526) |
| 0.1 |
0.5 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.1 |
0.9 |
GO:0007320 |
insemination(GO:0007320) |
| 0.1 |
0.4 |
GO:0060523 |
prostate epithelial cord elongation(GO:0060523) |
| 0.1 |
0.3 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.1 |
0.3 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 |
1.1 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
1.9 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
1.2 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 |
0.2 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 |
0.7 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.1 |
0.2 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 |
0.3 |
GO:0044830 |
modulation by host of viral RNA genome replication(GO:0044830) |
| 0.1 |
1.0 |
GO:1901629 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) regulation of presynaptic membrane organization(GO:1901629) |
| 0.1 |
0.2 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 |
0.2 |
GO:0090135 |
actin filament branching(GO:0090135) negative regulation of phospholipase C activity(GO:1900275) |
| 0.1 |
0.3 |
GO:0009098 |
branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 |
3.4 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.1 |
0.5 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
0.9 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.2 |
GO:0035878 |
nail development(GO:0035878) |
| 0.0 |
0.2 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 |
0.1 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 |
0.6 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.2 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.0 |
0.9 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.0 |
0.1 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.0 |
1.0 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
0.5 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.0 |
0.7 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.2 |
GO:0060414 |
aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 |
0.1 |
GO:0097475 |
motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 |
0.6 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.1 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.2 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 |
0.3 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.1 |
GO:2001137 |
positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) positive regulation of endocytic recycling(GO:2001137) |
| 0.0 |
0.8 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
0.5 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.1 |
GO:0045896 |
regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 |
0.4 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.0 |
0.4 |
GO:0045648 |
positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 |
0.9 |
GO:0048599 |
oocyte development(GO:0048599) |
| 0.0 |
0.4 |
GO:0030316 |
osteoclast differentiation(GO:0030316) |
| 0.0 |
0.2 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.2 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.1 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 |
0.5 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 |
0.1 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 |
0.1 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.1 |
GO:0000052 |
citrulline metabolic process(GO:0000052) |
| 0.0 |
0.3 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.2 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 |
0.4 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.0 |
GO:0009838 |
abscission(GO:0009838) |
| 0.0 |
0.2 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.3 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.0 |
0.3 |
GO:0003351 |
epithelial cilium movement(GO:0003351) |
| 0.0 |
0.4 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.0 |
0.7 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.5 |
GO:0021884 |
forebrain neuron development(GO:0021884) |
| 0.0 |
0.3 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.1 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) |
| 0.0 |
0.0 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.0 |
0.1 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.0 |
0.1 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 |
0.0 |
GO:1902512 |
positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |