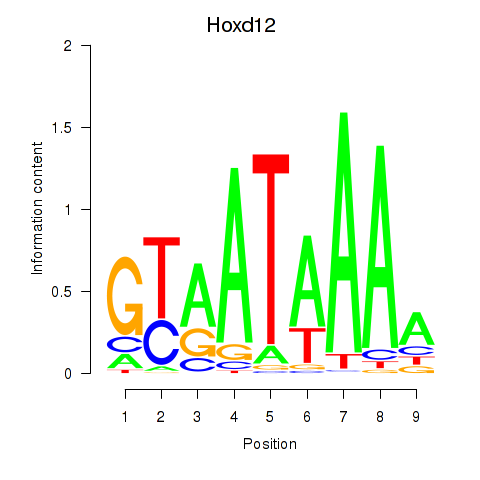
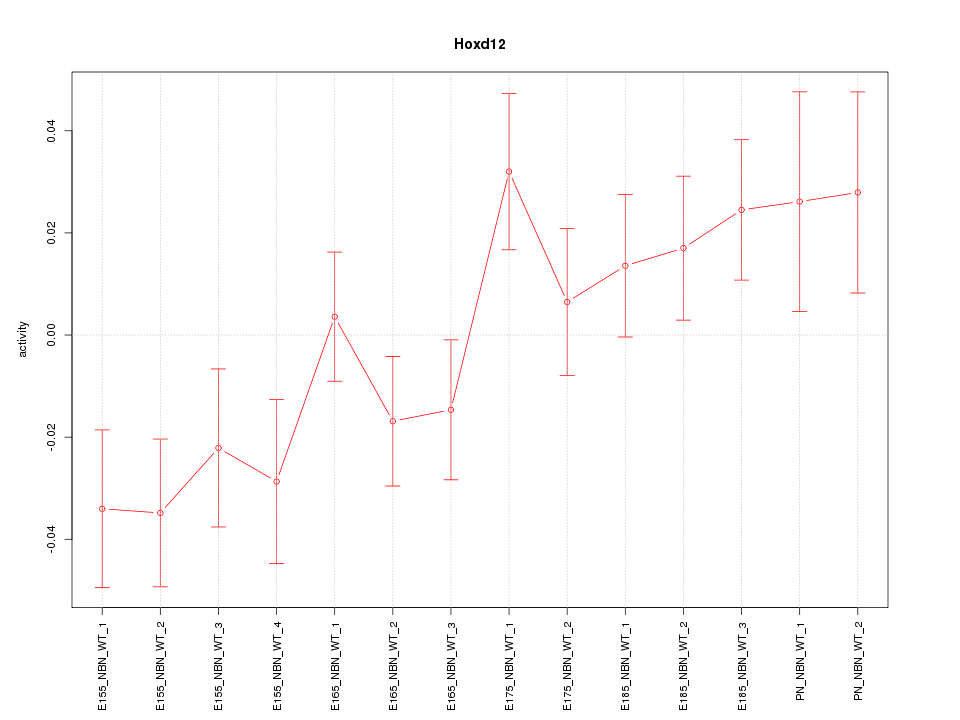
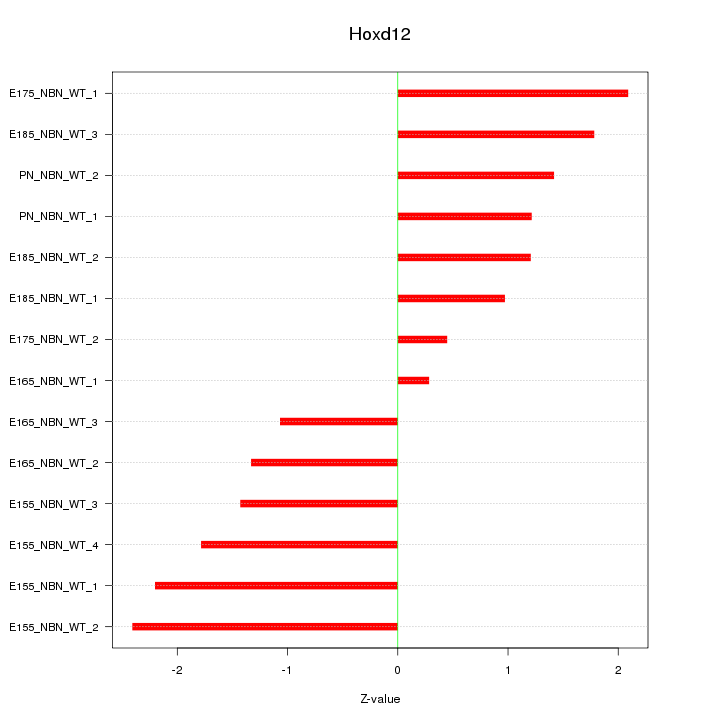
| 1.5 |
17.0 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 1.2 |
5.9 |
GO:2000325 |
regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.9 |
3.7 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.8 |
5.5 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.7 |
2.2 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.4 |
3.1 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.4 |
9.2 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.4 |
1.2 |
GO:0060423 |
limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) negative regulation of mitotic cell cycle, embryonic(GO:0045976) foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.4 |
1.1 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.4 |
1.1 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.3 |
2.4 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.3 |
1.4 |
GO:0090160 |
Golgi to lysosome transport(GO:0090160) |
| 0.3 |
1.9 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.2 |
1.7 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.2 |
0.7 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.2 |
3.6 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.2 |
1.1 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.2 |
1.6 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.2 |
3.2 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 |
0.7 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 |
5.9 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.7 |
GO:0007262 |
STAT protein import into nucleus(GO:0007262) |
| 0.1 |
0.4 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 |
0.4 |
GO:0060126 |
somatotropin secreting cell differentiation(GO:0060126) |
| 0.1 |
1.3 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.1 |
0.5 |
GO:0097118 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 |
1.0 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.1 |
3.5 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.1 |
1.7 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 |
0.6 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.1 |
0.4 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 |
2.1 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.1 |
0.3 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.1 |
0.5 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
0.3 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.1 |
0.9 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
1.7 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
0.7 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.1 |
2.4 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 |
0.3 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 |
0.4 |
GO:0097343 |
ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 |
0.2 |
GO:0060745 |
mammary gland branching involved in pregnancy(GO:0060745) |
| 0.1 |
2.1 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.1 |
1.2 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.1 |
0.1 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.8 |
GO:0055070 |
copper ion homeostasis(GO:0055070) |
| 0.0 |
1.6 |
GO:0010972 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 |
1.2 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.4 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.3 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
2.9 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
0.5 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 |
1.2 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.2 |
GO:0006527 |
arginine catabolic process(GO:0006527) |
| 0.0 |
1.5 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
2.7 |
GO:0030326 |
embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 |
0.7 |
GO:0045471 |
response to ethanol(GO:0045471) |
| 0.0 |
0.4 |
GO:0055010 |
ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.0 |
0.1 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |